Structural bases for CRMP function in plexin-dependent semaphorin3A signaling - PubMed (original) (raw)
Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Rahul C Deo et al. EMBO J. 2004.
Abstract
Collapsin response mediator proteins (CRMPs) are cytosolic phosphoproteins involved in neuronal differentiation and axonal guidance. CRMP2 was previously shown to mediate the repulsive effect of Sema3A on axons and to participate in axonal specification. The X-ray crystal structure of murine CRMP1 was determined at 2.1 A resolution and demonstrates that CRMP1 is a bilobed 'lung-shaped' protein forming a tetrameric assembly. Structure-based mutagenesis of surface-exposed residues was employed to map functional domains. As a rapid assay for CRMP, we exploited a reconstituted Sema3A signaling system in COS-7 cells expressing the receptor components Neuropilin1 and PlexinA1 (NP1/PlexA1). In these cells, CRMP and PlexA1 form a physical complex that is reduced in amount by NP1 but enhanced by Sema3A/NP1. Furthermore, CRMP accelerates Sema3A-induced cell contraction. Alanine substitutions in one domain of CRMP1 produce a constitutively active protein that causes Sema3A-independent COS-7 contraction. This mutant CRMP mimics the DRG neurite outgrowth-inhibiting effects of Sema3A and reduces Sema3A-induced axonal repulsion. These data provide a structural view of CRMP function in Plex-dependent Sema3A signaling.
Figures
Figure 1
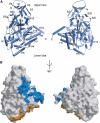
Three-dimensional structure of CRMP1 monomer. (A) BOBSCRIPT (Esnouf, 1999) drawing of the front and back views of CRMP1. α-Helices are labeled H1–H19 and β-strands are labeled S1–S21. The positions of the N- and C-termini are indicated. A thin line separates the N-terminal upper lobe from the C-terminal lower lobe. (B) GRASP (Nicholls et al, 1991) representations of the solvent-accessible surface of CRMP1 calculated using a water probe radius=1.4 Å. The surface overlying residues contributing to tetramer interfaces 1 and 2 are colored blue and gold, respectively. The orientations are identical to those in (A). Secondary structure elements contributing to the tetramer interfaces are labeled.
Figure 2
Sequence alignment of CRMPs and related proteins. Secondary structural elements were obtained from the X-ray structure. Gray circles denote disordered residues or residues not included in the construct (1–8, 526–572). Sequence homology is encoded by a gray → purple color gradient (40–100% identity). Functional classifications: 1, tetramer interface 1; 2, tetramer interface 2. Sequence numbering for mouse CRMP1 is provided in green above the secondary structure elements.
Figure 3
CRMP forms a
D
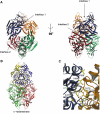
-hydantoinase-like tetramer. (A) BOBSCRIPT (Esnouf, 1999) drawing of CRMP1 tetramer. Individual protomers are colored blue, yellow, green, and red. The two intermolecular interfaces contributing to the tetramer are denoted by arrows. (B) MOLSCRIPT (Kraulis, 1991) drawing of
D
-hydantoinase tetramer. Individual protomers are colored blue, yellow, green, and red. (C) Detailed MOLSCRIPT (Kraulis, 1991) representation of a portion of CRMP-CRMP interface 1. Residues contributing to the interface are depicted as ball-and-stick figures, and hydrogen bonds are shown as dashed gray lines. Secondary structure elements are labeled in green and numbered according to Figure 1. Residues and secondary structure elements of the blue protomer are denoted with an apostrophe. Backbone atoms, which play a significant role in the H15–H16 loop interaction with helix H19′, have been omitted for clarity.
Figure 4
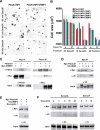
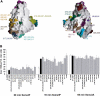
CRMPs accelerate the NP1/PlexA1-mediated Sema3A-induced COS7 cell contraction. (A) COS-7 cells expressing Myc-PlexA1/ NP1 alone or with CRMP1 were treated with AP-Sema3A or AP-Sema3F for the indicated times. Whereas Sema3F-treated cells do not exhibit any morphological changes, cells treated with Sema3A contract within 10 min in the presence of CRMP1. In contrast, cells expressing only PlexA1/NP1 shrink after 60 min Sema3A incubation. (B) Quantification of cellular areas (mean±s.e.m.) of COS-7 cells expressing PlexA1/NP1 and different CRMP isoforms and treated with Sema3F or Sema3A at 4 or 37°C. CRMPs 1–4 are designated as C1–4. All four CRMP isoforms were able to facilitate COS-7 contraction after Sema3A treatment for 10 min at 37°C. (C) HEK293T cells were transfected with vector or untagged full-length PlexA1 with Myc-tagged CRMP2, or vector or Myc-CRMP2 with PlexA1. Lysates were immunoprecipitated with anti-Myc or anti-Plex antibodies and immunoblotted with anti-Plex and anti-Myc antibodies. Note that PlexA1 co-precipitated with Myc-CRMP2 (left panel) and Myc-CRMP2 co-precipitated with anti-Plex (right panel). Input represents preimmune lysate. (D) HEK293T cells were transfected with vector or Myc-PlexA1 with V5-tagged CRMP1, CRMP3, or CRMP4. Lysates were immunoprecipitated with anti-Myc antibody, and the eluates were probed with anti-Myc or anti-V5 antibodies. All three CRMPs co-precipitate with Myc-PlexA1. (E) HEK293T cells were transfected with V5-CRMP4 and with either Myc-PlexA1 or NP1/Myc-PlexA1 dual-expression vector. Cell lysates were immunoprecipitated with anti-Myc antibody and blotted with anti-Myc and anti-V5 antibody. V5-CRMP4 co-precipitated with Myc-PlexA1 only in the absence of NP1. (F) V5-CRMP4- and NP1/Myc-PlexA1-transfected HEK293T cell lysates were treated with Sema3A or Sema3F at room temperature for the indicated periods of time. Lysates were immunoprecipitated with anti-Myc antibody and blotted with anti-Myc and anti-V5 antibodies. Co-precipitated V5-CRMP4 is detected after 5 min Sema3A treatment and can still be seen after 30 min Sema3A treatment. Sema3F does not increase the ability of Myc-PlexA1 to co-precipitate V5-CRMP4.
Figure 5
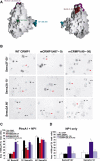
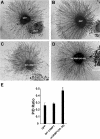
Effect of mCRMP1 mutants on COS-7 contraction. (A) GRASP (Nicholls et al, 1991) representation of CRMP1 monomer highlighting the surface regions affected by the four mCRMP1 mutants: mCRMP1(49–56) and mCRMP1(38,39,41,43) on the upper lobe, mCRMP1(367–368) in the central portion, and mCRMP1(487–489) on interface 1. (B) COS-7 cells expressing PlexA1 and NP1 with WT CRMP1, mCRMP1(487–489), or mCRMP1(49–56) were treated with Sema3A or Sema3F for the indicated times. The left column shows that cells expressing WT CRMP1 exhibit facilitated contraction after 10 min of Sema3A treatment but no contraction following 60 min with Sema3F. The middle column shows cells expressing mCRMP1(487–489) that fail to contract after 10 min Sema3A treatment, but are fully contracted after 60 min (similar to NP1/PlexA1 only expressing cells in Figure 1). The right column shows that cells expressing mCRMP1(49–56) are contracted in all treatment groups. Black arrows, black arrowheads, and red arrowheads indicate spread AP-stained cells, shrunk AP-stained cells, and unlabeled cells, respectively. (C) Quantification (mean±s.e.m.) of cell area of COS-7 cells expressing PlexA1 and NP1 with either WT CRMP1, or indicated mCRMP1 mutants. Mutant mCRMP1(487–489) and mCRMP1(367–368) failed to facilitate Sema3A-induced contraction, while mCRMP1(49–56) and mCRMP1(38,39,41,43) showed significant contraction with Sema3F treatment. (D) Quantification of cell area (mean±s.e.m.) of COS-7 cells expressing only NP1 and indicated CRMP1 construct, and treated with Sema3F for 60 min or Sema3A for 10 min. Cells expressing mCRMP1(49–56) were still contracted in the absence of PlexA1, while cells expressing mCRMP1(38,39,41,43) were spread (*P<0.005; **P<0.001).
Figure 6
Summary of mCRMP1 mutants that had no effect on Sema3A-induced COS-7 cell contraction. (A) GRASP (Nicholls et al, 1991) representation of CRMP1 monomer highlighting the sites of 12 mutations with phenotypes identical to WT CRMP1 in the COS-7 contraction assay. (B) Quantitation of cell area (mean±s.e.m.) of COS-7 cells expressing PlexA1, NP1, and indicated mCRMP1 constructs.
Figure 7
mCRMP1s are correctly localized in the cell and interact with both PlexA1 and CRMP2. (A) The subcellular localization of overexpressed mCRMP1s was examined in either transfected COS7 cells (upper row) or HSV-infected E8 chick DRG explants (lower row). All mCRMP1s showed a similar localization to overexpressed WT CRMP1. (B) HEK293T cells were transfected with either vector, V5-tagged WT CRMP1, or a V5-mCRMP1, and either Myc-tagged PlexA1 or CRMP2. Lysates were then immunoprecipitated with anti-V5 antibody and blotted with anti-Myc or anti-V5. Both Myc-PlexA1 and Myc-CRMP2 were able to co-immunoprecipitate with WT CRMP1 and all four mCRMP1s.
Figure 8
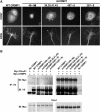
mCRMP1(49–56) reduces neurite outgrowth in DRG neurons. (A) Neurite outgrowth from dissociated E8 chick DRG neurons infected with HSV-WT CRMP1 or HSV-mCRMP1(49–56) is visualized by anti-V5 immunoflourescence. Cells were cultured with or without 100 nM Sema3A. (B) Total neurite length/cell (mean±s.e.m.) was measured for dissociated E8 chick DRG neurons infected with HSV-EGFP, or V5-tagged HSV-WT CRMP1 or HSV-mCRMP1(49–56). Neurons were cultured with or without 100 nM Sema3A. mCRMP1(49–56) significantly reduced outgrowth compared to EGFP or WT CRMP1 (**P<0.005); Sema3A reduced outgrowth only in EGFP and WT CRMP-1 cultures (*P<0.05).
Figure 9
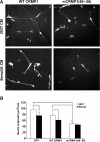
mCRMP1(49–56) attenuates Sema3A-mediated axon repulsion. (A–D) E8 chick DRG explants were infected with either HSV-GFP (A,B), or V5-tagged HSV-WT CRMP1 (C) or HSV-mCRMP1(49–56) (D), and grown in a collagen gel with Sema3A-expressing HEK293T cells (B,D) on control HEK293T (A) cells and visualized with GFP or V5 immunostaining. White dots represent the borders of axon growth. (E) Sema3A-mediated repulsion was quantitated by determining the ratio of axonal outgrowth proximal (P) and distal (D) to the Sema3A-expressing 293T cells (P/D ratio; mean±s.e.m.). Overexpression of mCRMP1(49–56) significantly reduced Sema3A-mediated axon repulsion (*P<0.005).
Similar articles
- Involvement of Fes/Fps tyrosine kinase in semaphorin3A signaling.
Mitsui N, Inatome R, Takahashi S, Goshima Y, Yamamura H, Yanagi S. Mitsui N, et al. EMBO J. 2002 Jul 1;21(13):3274-85. doi: 10.1093/emboj/cdf328. EMBO J. 2002. PMID: 12093729 Free PMC article. - The CRMP family of proteins and their role in Sema3A signaling.
Schmidt EF, Strittmatter SM. Schmidt EF, et al. Adv Exp Med Biol. 2007;600:1-11. doi: 10.1007/978-0-387-70956-7_1. Adv Exp Med Biol. 2007. PMID: 17607942 Free PMC article. Review. - Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein.
Schmidt EF, Shim SO, Strittmatter SM. Schmidt EF, et al. J Neurosci. 2008 Feb 27;28(9):2287-97. doi: 10.1523/JNEUROSCI.5646-07.2008. J Neurosci. 2008. PMID: 18305261 Free PMC article. - RanBPM contributes to Semaphorin3A signaling through plexin-A receptors.
Togashi H, Schmidt EF, Strittmatter SM. Togashi H, et al. J Neurosci. 2006 May 3;26(18):4961-9. doi: 10.1523/JNEUROSCI.0704-06.2006. J Neurosci. 2006. PMID: 16672672 Free PMC article. - Molecular basis of semaphorin-mediated axon guidance.
Nakamura F, Kalb RG, Strittmatter SM. Nakamura F, et al. J Neurobiol. 2000 Aug;44(2):219-29. doi: 10.1002/1097-4695(200008)44:2<219::aid-neu11>3.0.co;2-w. J Neurobiol. 2000. PMID: 10934324 Review.
Cited by
- Spastin interacts with collapsin response mediator protein 3 to regulate neurite growth and branching.
Ji ZS, Li JP, Fu CH, Luo JX, Yang H, Zhang GW, Wu W, Lin HS. Ji ZS, et al. Neural Regen Res. 2021 Dec;16(12):2549-2556. doi: 10.4103/1673-5374.313052. Neural Regen Res. 2021. PMID: 33907047 Free PMC article. - Structure of human collapsin response mediator protein 1: a possible role of its C-terminal tail.
Liu SH, Huang SF, Hsu YL, Pan SH, Chen YJ, Lin YH. Liu SH, et al. Acta Crystallogr F Struct Biol Commun. 2015 Aug;71(Pt 8):938-45. doi: 10.1107/S2053230X15009243. Epub 2015 Jul 28. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26249678 Free PMC article. - Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones.
Arimura N, Ménager C, Kawano Y, Yoshimura T, Kawabata S, Hattori A, Fukata Y, Amano M, Goshima Y, Inagaki M, Morone N, Usukura J, Kaibuchi K. Arimura N, et al. Mol Cell Biol. 2005 Nov;25(22):9973-84. doi: 10.1128/MCB.25.22.9973-9984.2005. Mol Cell Biol. 2005. PMID: 16260611 Free PMC article. - Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications.
Hensley K, Venkova K, Christov A, Gunning W, Park J. Hensley K, et al. Mol Neurobiol. 2011 Jun;43(3):180-91. doi: 10.1007/s12035-011-8166-4. Epub 2011 Jan 28. Mol Neurobiol. 2011. PMID: 21271304 Review. - Feature amplified voting algorithm for functional analysis of protein superfamily.
Hung CL, Lee C, Lin CY, Chang CH, Chung YC, Yi Tang C. Hung CL, et al. BMC Genomics. 2010 Dec 1;11 Suppl 3(Suppl 3):S14. doi: 10.1186/1471-2164-11-S3-S14. BMC Genomics. 2010. PMID: 21143781 Free PMC article.
References
- Abendroth J, Niefind K, May O, Siemann M, Syldatk C, Schomburg D (2002a) The structure of L-hydantoinase from Arthobacter aurescens leads to an understanding of dihydropyrimidinase substrate and enantio specificity. Biochemistry 41: 8589–8597 - PubMed
- Abendroth J, Niefind K, Schomburg D (2002b) X-ray structure of a dihydropyrimidinase from Thermus sp. at 1.3 A resolution. J Mol Biol 320: 143–156 - PubMed
- Aizawa H, Wakatsuki S, Ishii A, Moriyama K, Sasaki Y, Ohashi K, Sekine-Aizawa Y, Sehara-Fujisawa A, Mizuno K, Goshima Y, Yahara I (2001) Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci 4: 367–373 - PubMed
- Benini S, Ciurli S, Nolting HF, Mangani S (1996) X-ray absorption spectroscopy study of native and phenylphosphorodiamidate-inhibited Bacillus pasteurii urease. Eur J Biochem 239: 61–66 - PubMed
- Benning MM, Kuo JM, Raushel FM, Holden HM (1995) Three-dimensional structure of the binuclear metal center of phosphotriesterase. Biochemistry 34: 7973–7978 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous