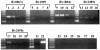Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo - PubMed (original) (raw)
Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo
JoAnn M Tufariello et al. Infect Immun. 2004 Jan.
Abstract
Mycobacterium tuberculosis possesses five genes with significant homology to the resuscitation-promoting factor (Rpf) of Micrococcus luteus. The M. luteus Rpf is a secreted approximately 16-kDa protein which restores active growth to cultures of M. luteus rendered dormant by prolonged incubation in stationary phase. More recently, the Rpf-like proteins of M. tuberculosis have been shown to stimulate the growth of extended-stationary-phase cultures of Mycobacterium bovis BCG. These data suggest that the Rpf proteins can influence the growth of mycobacteria; however, the studies do not demonstrate specific functions for the various members of this protein family, nor do they assess the function of M. tuberculosis Rpf homologues in vivo. To address these questions, we have disrupted each of the five rpf-like genes in M. tuberculosis Erdman, and analyzed the mutants for their growth in vitro and in vivo. In contrast to M. luteus, for which rpf is an essential gene, we find that all of the M. tuberculosis rpf deletion mutant strains are viable; in addition, all show growth kinetics similar to Erdman wild type both in vitro and in mouse organs following aerosol infection. Analysis of rpf expression in M. tuberculosis cultures from early log phase through late stationary phase indicates that expression of the rpf-like genes is growth phase-dependent, and that the expression patterns of the five M. tuberculosis rpf genes, while overlapping to various degrees, are not uniform. We also provide evidence that mycobacterial rpf genes are expressed in vivo in the lungs of mice acutely infected with virulent M. tuberculosis.
Figures
FIG. 1.
Construction of rpf deletion mutants by allelic replacement. Shown in each panel is a schematic illustrating the chromosomal organization of the wild-type (upper) and rpf deletion mutant (lower) strains for Rv0867c (A), Rv1009 (B), Rv1884c (C), Rv2389c (D), and Rv2450c (E), in which each of the individual rpf_-like genes has been disrupted by a hygromycin resistance cassette. The schematics also indicatethe restriction enzymes used to digest the genomic DNA for Southern blot analysis and the sizes of expected hybridizing bands for each strain (indicated by the dashed lines). Below each diagram of the chromosome is a Southern blot showing digested genomic DNA for several independent clones of the Δ_rpf mutant (numbered) and for wild-type Erdman (always the lane on the far right [WT]) probed with the flanking region of the gene. The migration of molecular size markers (in kilobases) is indicated to the left of each blot. The location of each probe is indicated in the schematic above. Below the ΔRv1009 diagram in panel B are two Southern blots: on the left is mc23126 (ΔRv1009) and on the right is mc23126A (ΔRv1009 rev).
FIG. 2.
Southern blot analysis of complementation of ΔRv1009. Shown are Southern blots of the complemented strains for mc23126 (ΔRv1009) and mc23126A (ΔRv1009 rev), each probed with the coding sequence of the complementing gene, and each panel showing (i) the original knockout strain (KO) in the far left lane, (ii) several independent complemented clones (numbered) in the middle lanes, and (iii) Erdman wild type (WT) in the far right lane. In all panels the migration of molecular size markers (in kilobases) is indicated at left. (A) For analysis of mc23301(ΔRv1009 complemented with Rv1009), chromosomal DNA was digested with _Eco_RI and probed with the Rv1009 coding sequence (primer pair 52/53 PCR product), yielding no hybridizing band in the original knockout strain, a 7,340-bp band in the complemented strains (lanes 1 to 5) and an 8,730-bp band for Erdman WT. (B) For analysis of mc23301A (ΔRv1009 rev complemented with Rv1009), chromosomal DNA was digested as in panel A above, the probe was again the Rv1009 coding sequence, and the expected band sizes are as noted in panel A above. In this panel the complemented strains are in lanes 1 to 4. (C) For analysis of mc23302 (ΔRv1009 complemented with ksgA), genomic DNA was digested with _Eco_RI and probed with the ksgA coding sequence (primer pair 60-61 PCR product), yielding a single band of 8,730 bp for Erdman WT, a single band of 1,960 bp for the original knockout strain (ΔRv1009), and two bands of 7,223 and 1,960 bp for the complemented strains (lanes 1 to 5). The complemented strains have two hybridizing bands because nearly the entire ksgA coding sequence was left intact in this knockout; the complementation was performed due to concerns about polar effects on ksgA expression. Therefore, each complemented strain has one band which comigrates with the band present in the original knockout strain and one additional band corresponding to insertion of a second copy of the ksgA gene at the attB site. (D) For analysis of mc23302A (ΔRv1009 rev complemented with ksgA), genomic DNA was digested and probed as above in panel C and the expected band sizes are as noted above; here the complemented strains are in lanes 1 to 4.
FIG. 3.
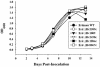
In vitro growth kinetics of M. tuberculosis Erdman wild type and the five rpf deletion mutant strains in Middlebrook 7H9 medium at 37°C with agitation. The ΔRv1009 strain used in this experiment was mc23126. We also found that strain mc23126A (ΔRv1009 rev) grew in a manner indistinguishable from wild type in 7H9 medium and that the number of CFU at each time point was similar for each of the strains tested (not shown).
FIG. 4.
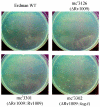
Colony size of the original ΔRv1009 strain (mc23126) compared with Erdman wild type and the effect on colony size of complementation with Rv1009 and with ksgA. Cultures were grown and transferred onto plates with Middlebrook 7H10 agar, and colonies were photographed at 23 days postinoculation as described in Materials and Methods. The strains in each panel are indicated. There were similar numbers of colonies on each plate.
FIG. 5.
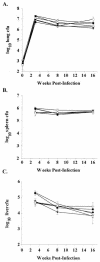
Disruption of individual _rpf_-like genes does not alter bacterial burden in organs of C57BL/6 mice through 16 weeks postinfection. Bacterial numbers in the lung (A), spleen (B), and liver (C) of C57BL/6 mice infected by aerosol with either Erdman wild type (WT) (•), mc23125 (ΔRv2389c) (○), mc23126A (ΔRv1009rev) (▾), mc23127 (ΔRv2450c) (▿), mc23128 (ΔRv1884c) (▪), or mc23129 (ΔRv0867c) (□) were determined. Data (means ± standard error [error bars] of three to four mice per group) are representative of three independent experiments.
FIG. 6.
Expression of the _M. tuberculosis rpf_-like genes in vitro as assessed by quantitative dilutional PCR. RNA was prepared from Erdman grown in 7H9 medium, and 1 μg of total RNA was reverse transcribed and serially diluted fivefold for use as template in PCRs to amplify each of the _rpf_-like genes. Primers, as described in Materials and Methods, were used to amplify portions of Rv0867c (A), Rv1009 (B), Rv1884c (C), Rv2389c (D), and Rv2450c (E). (A to E) Lanes 1 to 5 contain PCR products of serial dilutions of cDNA from day 4 (OD600 = 0.125), lanes 6 to 10 are from day 6 (OD600 = 0.574), lanes 11 to 15 are from day 8 (OD600 = 1.408), and lanes 16 to 20 are from day 12 (OD600 = 2.512). (F) PCR products of cDNAs from day 4 (lanes 1 and 5), day 6 (lanes 2 and 6), day 8 (lanes 3 and 7), and day 12 (lanes 4 and 8) using 16S rRNA primers (lanes 1 to 4, 9, and 10) and 23S rRNA primers (lanes 5 to 8, 11, and 12). Lanes 9 and 11 contain no template (negative controls), while lanes 10 and 12 contain Erdman genomic DNA as the template. Lanes 16 to 19 show that no PCR product was obtained from non-reverse-transcribed RNA from day 4 (lane16), day 6 (lane 17), day 8 (lane 18), or day 12 (lane 19) cultures, using primers to amplify Rv1884c. A 50-bp ladder (Invitrogen) is shown in the far left lane of each gel.
FIG. 7.
Expression of M. tuberculosis rpf homologues in a late-stationary-phase (4-month-old) Erdman culture. RNA was prepared from Erdman grown in 7H9 medium, and 5 μg of total RNA was reverse transcribed and serially diluted fivefold for use as template in PCRs to amplify each of the _rpf_-like genes. Primers, as described in Materials and Methods, were used to amplify portions of Rv0867c (lanes 1 to 4), Rv1009 (lanes 5 to 8), Rv1884c (lanes 9 to 12), Rv2389c (lanes 13 to 16), and Rv2450c (lanes 17 to 20). Lane 21 is a negative control containing no template, and lane 22 shows amplification of 23S rRNA from the same cDNA template used to amplify the rpf genes. Lanes 23 to 28 contain non-reverse-transcribed RNA as the template, using primers to amplify Rv2389c (lane 23), Rv1009 (lane 24), Rv2450c (lane 25), Rv1884c (lane 26), Rv0867c (lane 27), and 23S rRNA (lane 28). A 50-bp ladder (Invitrogen) is shown to the left of each series of samples.
FIG. 8.
In vivo expression of _M. tuberculosis rpf_-like genes. RNA was prepared from lungs of mice infected with a clinical strain of M. tuberculosis, and total RNA (equivalent of 1.25 μg per PCR mixture) was reverse transcribed and used as the template in PCRs to amplify each of the _rpf_-like genes or to amplify 23S rRNA, as indicated by the gene designation above each lane. Also shown are the results of PCR amplification using RNA which had not been reverse transcribed (No RT), employing 23S rRNA primers, for both lung 1 and lung 2. The far right lane of each gel shows the results of PCR amplification using 23S rRNA primers with water (H2O) as template.
Similar articles
- Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro.
Downing KJ, Mischenko VV, Shleeva MO, Young DI, Young M, Kaprelyants AS, Apt AS, Mizrahi V. Downing KJ, et al. Infect Immun. 2005 May;73(5):3038-43. doi: 10.1128/IAI.73.5.3038-3043.2005. Infect Immun. 2005. PMID: 15845511 Free PMC article. - Deletion of the Mycobacterium tuberculosis resuscitation-promoting factor Rv1009 gene results in delayed reactivation from chronic tuberculosis.
Tufariello JM, Mi K, Xu J, Manabe YC, Kesavan AK, Drumm J, Tanaka K, Jacobs WR Jr, Chan J. Tufariello JM, et al. Infect Immun. 2006 May;74(5):2985-95. doi: 10.1128/IAI.74.5.2985-2995.2006. Infect Immun. 2006. PMID: 16622237 Free PMC article. - A family of autocrine growth factors in Mycobacterium tuberculosis.
Mukamolova GV, Turapov OA, Young DI, Kaprelyants AS, Kell DB, Young M. Mukamolova GV, et al. Mol Microbiol. 2002 Nov;46(3):623-35. doi: 10.1046/j.1365-2958.2002.03184.x. Mol Microbiol. 2002. PMID: 12410821 - Rpf Proteins Are the Factors of Reactivation of the Dormant Forms of Actinobacteria.
Nikitushkin VD, Demina GR, Kaprelyants AS. Nikitushkin VD, et al. Biochemistry (Mosc). 2016 Dec;81(13):1719-1734. doi: 10.1134/S0006297916130095. Biochemistry (Mosc). 2016. PMID: 28260493 Review. - Resuscitation-promoting factors are important determinants of the pathophysiology in Mycobacterium tuberculosis infection.
Rosser A, Stover C, Pareek M, Mukamolova GV. Rosser A, et al. Crit Rev Microbiol. 2017 Sep;43(5):621-630. doi: 10.1080/1040841X.2017.1283485. Epub 2017 Feb 17. Crit Rev Microbiol. 2017. PMID: 28338360 Review.
Cited by
- Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro.
Downing KJ, Mischenko VV, Shleeva MO, Young DI, Young M, Kaprelyants AS, Apt AS, Mizrahi V. Downing KJ, et al. Infect Immun. 2005 May;73(5):3038-43. doi: 10.1128/IAI.73.5.3038-3043.2005. Infect Immun. 2005. PMID: 15845511 Free PMC article. - Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro.
Hingley-Wilson SM, Lougheed KE, Ferguson K, Leiva S, Williams HD. Hingley-Wilson SM, et al. Tuberculosis (Edinb). 2010 Jul;90(4):236-44. doi: 10.1016/j.tube.2010.03.013. Epub 2010 Jun 11. Tuberculosis (Edinb). 2010. PMID: 20541977 Free PMC article. - A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes.
Russell-Goldman E, Xu J, Wang X, Chan J, Tufariello JM. Russell-Goldman E, et al. Infect Immun. 2008 Sep;76(9):4269-81. doi: 10.1128/IAI.01735-07. Epub 2008 Jun 30. Infect Immun. 2008. PMID: 18591237 Free PMC article. - Expression of Resuscitation-Promoting Factor C Stimulates the Growth of Mycobacterium bovis BCG and Delays DevR Regulon Activation in Hypoxia.
Reyes-González LV, Hernández de la Cruz ON, Castañón-Arreola M. Reyes-González LV, et al. Int J Microbiol. 2025 Feb 17;2025:2139933. doi: 10.1155/ijm/2139933. eCollection 2025. Int J Microbiol. 2025. PMID: 39996126 Free PMC article. - Genomic analysis of Mycobacterium tuberculosis variant bovis strains isolated from bovine in the state of Mato Grosso, Brazil.
Dos Anjos TR, Castro VS, Machado Filho ES, Suffys PN, Gomes HM, Duarte RS, Figueiredo EES, Carvalho RCT. Dos Anjos TR, et al. Front Vet Sci. 2022 Nov 16;9:1006090. doi: 10.3389/fvets.2022.1006090. eCollection 2022. Front Vet Sci. 2022. PMID: 36467663 Free PMC article.
References
- Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. - PubMed
- Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAMA 282:677-686. - PubMed
- Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI026170/AI/NIAID NIH HHS/United States
- AI49375/AI/NIAID NIH HHS/United States
- AI36990/AI/NIAID NIH HHS/United States
- R01 AI026170/AI/NIAID NIH HHS/United States
- K08 AI049375/AI/NIAID NIH HHS/United States
- AI26170/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases