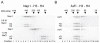Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities - PubMed (original) (raw)
Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities
Michael Bruno et al. Mol Cell. 2003 Dec.
Abstract
ATP-dependent chromatin remodeling activities function to manipulate chromatin structure during gene regulation. One of the ways in which they do this is by altering the positions of nucleosomes along DNA. Here we provide support for the ability of these complexes to move nucleosomes into positions in which DNA is unraveled from one edge. This is expected to result in the loss of histone-DNA contacts that are important for retention of one H2A/H2B dimer within the nucleosome. Consistent with this we find that several chromatin remodeling complexes are capable of catalyzing the exchange of H2A/H2B dimers between chromatin fragments in an ATP-dependent reaction. This provides eukaryotes with additional means by which they may manipulate chromatin structure.
Figures
Figure 1
Histone binding to Nap1, but not Asf1, shows sensitivity to ionic strength. (A) The elution profiles of Nap1-H3-H4 from gel filtration shown as consecutive fractions separated by SDS-PAGE. Complex formation under 0.6 M NaCl and 1.0 M NaCl was monitored. Fraction numbers are denoted at the bottom of the gel and the elution points of molecular weight markers shown above. (B) The same experiment carried out for Asf1 in complex with H3 and H4.
Figure 2
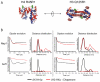
Directed sulfhydryl reactive crosslinking suggests Nap1 binds H3 and H4 in their tetrameric conformation. (A) The structure of the (H3-H4)2-tetramer from the nucleosome crystal structure showing the proximity of H3 K115 residues (green spheres) (image created using Pymol,
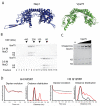
). Chemical structure of the compound MTS-3-MTS used to crosslink cysteines engineered at position 115 is shown alongside. (B) The extent of crosslinking under increasing concentrations of cysteine free Nap1 or Asf1 shown by separation of the reaction components by SDS-PAGE. Concentration of chaperone from left to right was 0.25, 0.5, 1.0, 2.0 μM monomer. (C) Gel filtration analysis of the H3 K115C crosslinked tetramer in the presence of Nap1 showing they remain stably associated under the conditions used. (D) Globular Asf1 was mixed with non-crosslinked and crosslinked tetramer and the complexes separated by gel filtration chromatography. The majority of Asf1 does not form a complex with crosslinked tetramers.
Figure 3
Extraction of long-range distances between spin labelled H3-H4 dimers agrees with a tetrameric conformation when bound to Nap1. (A) Positions of spin labels on H4 R45 and H3 Q125 (green spheres) are shown on the structure of the tetramer taken from the nucleosome (PDB code 1KX5, images created by Pymol,
). (B) The background corrected dipolar evolution and the distance distribution for each labelling site in complex with Nap1 and Asf1 is shown (red trace). For comparison the tetramer value extracted from the tetramer alone is shown alongside (black trace).
Figure 4
Vps75 binds H3 and H4 in their tetrameric conformation. (A) Nap1 (PDB code: 2ZR2) and Vps75 (PDB code: 3C9D) both adopt the headphone fold. Structures shown for comparison (images created by Pymol,
). (B) H3 and H4 binding by Vps75 displays sensitivity to ionic strength. Fractions from gel filtration chromatography are shown under 0.4 M and 0.6 M sodium chloride. Vps75-H3-H4 form a complex at 0.4 M sodium chloride (fractions 7-9, top panel), but elute in their separate fractions at 0.6 M (note, both free Vps75 dimer and H3-H4 tetramer elute in fractions 13-15) (C) Histone tetramer specific crosslinking of H3 K115C by MTS-3-MTS persists in the presence of cysteine free Vps75. Concentration of Vps75 from left to right was 0.25, 0.5, 1.0, 2.0 μM monomer. (D) Long range distance from histone tetramer spin labelled at position H4 R45R1 and H3 Q125R1 complexed with Vps75 were extracted using the PELDOR experiment. Background correct dipolar evolutions and distance distributions for each are shown (red trace). For comparison, distances measured for the tetramer alone are shown alongside (black trace).
Figure 5
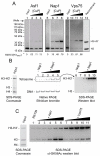
Nap1 and Vps75 co-purify with tetrameric H3-H4 and can use H3 and H4 trapped in their tetrameric conformation for their biological functions in vitro. (A)TAP-tagged Asf1, Nap1 and Vps75 were purified from strains expressing H3K115C-myc as the sole source of histone H3. Complexes were subject to crosslinking in the presence of 0.064, 0.25 or 1.0 mM CuP (Asf1, lanes 2-4; Nap1, lanes 6-8; Vps75, lanes 10-12), separated by SDS-PAGE and immuno-blotted within an anti-myc antibody. The intensity of the anti-myc signal was quantified and the proportion of crosslinked H3 of the total displayed at the bottom of each lane. Lane 13 contains recombinant partially cross-linked (non-tagged) H3 developed with a H3 antibody as a size standard (B) (H3-H4)2-tetramer deposition by Nap1. Left panel: crosslinked tetramer input stained with coomassie. This indicates that the majority of histone H3 present is crosslinked. Centre panel: monotetrasomes were separated from free DNA by native gel electrophoresis. Lane 1, without Nap1; lane 2, 3, 4 and 5: 0.2, 0.5, 1.0 and 2.0 μM Nap1, respectively. Right panel: the mono-tetrasomal bands were excised from the native gel and subject to SDS-PAGE and probed by western blotting for H3. Lanes 6-10 correspond to lanes 1-5 from the centre panel. This shows that crosslinked tetramers are present in tetrasomes assembled by Nap1. (C) Vps75 preferentially promotes Rtt109 acetylation of crosslinked tetramer, whereas Asf1 preferentially directs Rtt109 acetylation in favour of non-crosslinked tetramer. Left panel: equimolar amounts of crosslinked to non-crosslinked H3 K115C was used as an input. Right panel: lane 1, Rtt109 alone (140 nM). Lanes 2-6: 60, 80, 100, 120 and 140 nM each of Asf1 and Rtt109, respectively. Lanes 7-11: 10, 15, 20, 25 and 30 nM of (Vps75)2-Rtt109 complex, respectively.
Similar articles
- p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone.
Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. Ito T, et al. Genes Dev. 2000 Aug 1;14(15):1899-907. Genes Dev. 2000. PMID: 10921904 Free PMC article. - Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP. Eustermann S, et al. Nature. 2018 Apr;556(7701):386-390. doi: 10.1038/s41586-018-0029-y. Epub 2018 Apr 11. Nature. 2018. PMID: 29643509 Free PMC article. - NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex.
Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Côté J. Altaf M, et al. J Biol Chem. 2010 May 21;285(21):15966-77. doi: 10.1074/jbc.M110.117069. Epub 2010 Mar 23. J Biol Chem. 2010. PMID: 20332092 Free PMC article. - Chromatin remodeling factors and DNA replication.
Varga-Weisz P. Varga-Weisz P. Prog Mol Subcell Biol. 2005;38:1-30. doi: 10.1007/3-540-27310-7_1. Prog Mol Subcell Biol. 2005. PMID: 15881889 Review. - ATP-dependent nucleosome remodeling.
Becker PB, Hörz W. Becker PB, et al. Annu Rev Biochem. 2002;71:247-73. doi: 10.1146/annurev.biochem.71.110601.135400. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045097 Review.
Cited by
- Inducible nucleosome depletion at OREBP-binding-sites by hypertonic stress.
Tong EH, Guo JJ, Xu SX, Mak K, Chung SK, Chung SS, Huang AL, Ko BC. Tong EH, et al. PLoS One. 2009 Dec 24;4(12):e8435. doi: 10.1371/journal.pone.0008435. PLoS One. 2009. PMID: 20041176 Free PMC article. - Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability.
Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Ferreira H, et al. Mol Cell Biol. 2007 Jun;27(11):4037-48. doi: 10.1128/MCB.02229-06. Epub 2007 Mar 26. Mol Cell Biol. 2007. PMID: 17387148 Free PMC article. - Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes.
Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongélard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, Bouvet P. Angelov D, et al. EMBO J. 2006 Apr 19;25(8):1669-79. doi: 10.1038/sj.emboj.7601046. Epub 2006 Apr 6. EMBO J. 2006. PMID: 16601700 Free PMC article. - Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer.
Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. Shukla MS, et al. Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1936-41. doi: 10.1073/pnas.0904497107. Epub 2010 Jan 13. Proc Natl Acad Sci U S A. 2010. PMID: 20080697 Free PMC article. - Nucleosomes can invade DNA territories occupied by their neighbors.
Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Engeholm M, et al. Nat Struct Mol Biol. 2009 Feb;16(2):151-8. doi: 10.1038/nsmb.1551. Epub 2009 Feb 1. Nat Struct Mol Biol. 2009. PMID: 19182801 Free PMC article.
References
- Annunziato AT. Split decision: What happens to nucleosomes during DNA replication? Journal of Biological Chemistry. 2005;280:12065–12068. - PubMed
- Baxevanis AD, Godfrey JE, Moudrianakis EN. Associative behavior of the histone (H3-H4)2 Tetramer: Dependence on ionic environment. Biochemistry. 1991;30:8817–8823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases