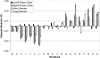Protein flexibility and intrinsic disorder - PubMed (original) (raw)
Comparative Study
Protein flexibility and intrinsic disorder
Predrag Radivojac et al. Protein Sci. 2004 Jan.
Abstract
Comparisons were made among four categories of protein flexibility: (1) low-B-factor ordered regions, (2) high-B-factor ordered regions, (3) short disordered regions, and (4) long disordered regions. Amino acid compositions of the four categories were found to be significantly different from each other, with high-B-factor ordered and short disordered regions being the most similar pair. The high-B-factor (flexible) ordered regions are characterized by a higher average flexibility index, higher average hydrophilicity, higher average absolute net charge, and higher total charge than disordered regions. The low-B-factor regions are significantly enriched in hydrophobic residues and depleted in the total number of charged residues compared to the other three categories. We examined the predictability of the high-B-factor regions and developed a predictor that discriminates between regions of low and high B-factors. This predictor achieved an accuracy of 70% and a correlation of 0.43 with experimental data, outperforming the 64% accuracy and 0.32 correlation of predictors based solely on flexibility indices. To further clarify the differences between short disordered regions and ordered regions, a predictor of short disordered regions was developed. Its relatively high accuracy of 81% indicates considerable differences between ordered and disordered regions. The distinctive amino acid biases of high-B-factor ordered regions, short disordered regions, and long disordered regions indicate that the sequence determinants for these flexibility categories differ from one another, whereas the significantly-greater-than-chance predictability of these categories from sequence suggest that flexible ordered regions, short disorder, and long disorder are, to a significant degree, encoded at the primary structure level.
Figures
Figure 1.
Amino acid compositions of various data sets. The composition of each amino acid of a reference data set of ordered proteins, Globular-3D, is subtracted from the composition of the four sets described herein; thus, negative peaks indicate depletions compared to the ordered reference set, and positive peaks represent enrichments. The order of the amino acids along the x-axis is from the most buried (left) to the most exposed (right) in typical globular proteins. Error bars indicate one standard deviation. Methionine at the N terminus and His-tags were not included in calculations.
Similar articles
- Predicting intrinsic disorder from amino acid sequence.
Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Obradovic Z, et al. Proteins. 2003;53 Suppl 6:566-72. doi: 10.1002/prot.10532. Proteins. 2003. PMID: 14579347 - Bioinformatical parsing of folding-on-binding proteins reveals their compositional and evolutionary sequence design.
Narasumani M, Harrison PM. Narasumani M, et al. Sci Rep. 2015 Dec 18;5:18586. doi: 10.1038/srep18586. Sci Rep. 2015. PMID: 26678310 Free PMC article. - Intrinsic disorder in the Protein Data Bank.
Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Le Gall T, et al. J Biomol Struct Dyn. 2007 Feb;24(4):325-42. doi: 10.1080/07391102.2007.10507123. J Biomol Struct Dyn. 2007. PMID: 17206849 - Intrinsically disordered protein.
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Dunker AK, et al. J Mol Graph Model. 2001;19(1):26-59. doi: 10.1016/s1093-3263(00)00138-8. J Mol Graph Model. 2001. PMID: 11381529 Review. - Evolution and disorder.
Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Brown CJ, et al. Curr Opin Struct Biol. 2011 Jun;21(3):441-6. doi: 10.1016/j.sbi.2011.02.005. Epub 2011 Apr 7. Curr Opin Struct Biol. 2011. PMID: 21482101 Free PMC article. Review.
Cited by
- Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines.
Vullo A, Bortolami O, Pollastri G, Tosatto SC. Vullo A, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W164-8. doi: 10.1093/nar/gkl166. Nucleic Acids Res. 2006. PMID: 16844983 Free PMC article. - Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources.
Mizianty MJ, Stach W, Chen K, Kedarisetti KD, Disfani FM, Kurgan L. Mizianty MJ, et al. Bioinformatics. 2010 Sep 15;26(18):i489-96. doi: 10.1093/bioinformatics/btq373. Bioinformatics. 2010. PMID: 20823312 Free PMC article. - Melatonin: Regulation of Viral Phase Separation and Epitranscriptomics in Post-Acute Sequelae of COVID-19.
Loh D, Reiter RJ. Loh D, et al. Int J Mol Sci. 2022 Jul 23;23(15):8122. doi: 10.3390/ijms23158122. Int J Mol Sci. 2022. PMID: 35897696 Free PMC article. Review. - Evaluation of disorder predictions in CASP9.
Monastyrskyy B, Fidelis K, Moult J, Tramontano A, Kryshtafovych A. Monastyrskyy B, et al. Proteins. 2011;79 Suppl 10(S10):107-18. doi: 10.1002/prot.23161. Epub 2011 Sep 16. Proteins. 2011. PMID: 21928402 Free PMC article. - Enhancing the activity of Bacillus circulans xylanase by modulating the flexibility of the hinge region.
Kazuyo F, Hong SY, Yeon YJ, Joo JC, Yoo YJ. Kazuyo F, et al. J Ind Microbiol Biotechnol. 2014 Aug;41(8):1181-90. doi: 10.1007/s10295-014-1454-z. Epub 2014 May 22. J Ind Microbiol Biotechnol. 2014. PMID: 24849049
References
- Altman, R.B., Hughes, C., and Jardetzky, O. 1994. Compositional characteristics of disordered regions in proteins. Prot. Pept. Lett. 2 120–127.
- Benner, S.A., Cohen, M.A., and Gerloff, D. 1992. Correct structure prediction? Nature 359 781. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources