Identification of many microRNAs that copurify with polyribosomes in mammalian neurons - PubMed (original) (raw)
Identification of many microRNAs that copurify with polyribosomes in mammalian neurons
John Kim et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Localized translation in mammalian dendrites may play a role in synaptic plasticity and contribute to the molecular basis for learning and memory. The regulatory mechanisms that control localized translation in neurons are not well understood. We propose a role for microRNAs (miRNAs), a class of noncoding RNAs, as mediators of neuronal translational regulation. We have identified 86 miRNAs expressed in mammalian neurons, of which 40 have not previously been reported. A subset of these miRNAs exhibits temporally regulated expression in cortical cultures. Moreover, all of the miRNAs that were tested cofractionate with polyribosomes, the sites of active translation. These findings indicate that a large, diverse population of miRNAs may function to regulate translation in mammalian neurons.
Figures
Fig. 1.
miRNAs from rat cortical neurons cluster in miRNA “families.” Previously identified miRNA sequences and miRNAs cloned in this study from combined E18 rat cortical neurons cultured for 1.5, 7, and 14 days were compiled and each sequence was aligned against all others by using a Smith-Waterman algorithm (EMBOSS 2.3.1; ref. 70). Complete hierarchical clustering was performed on the dissimilarity matrix generated from the scores of the pairwise sequence alignments. A dendrogram was generated from the clustering analysis (see Fig. 5) and was cut to yield a set of clusters, which were hand-adjusted to improve groupings of miRNAs that share common subsequences. Two examples of such clusters are presented here. Conserved sequences are highlighted in yellow. miRNAs identified in rat cortical neurons are labeled in red. R.n., Rattus norvegicus; M.m., Mus musculus; H.s., Homo sapiens; C.e., Caenorhabditis elegans. H/M/R designates clones found in human, mouse, and rat.
Fig. 2.
Tissue-specific expression patterns of miRNAs cloned from rat cortex. Northern blots of total RNA isolated from adult rat tissues were assayed with probes against 12 miRNAs. Eight miRNAs were expressed predominantly in the cortex and cerebellum. Two miRNAs are shown in a: _mir_-_129_-2* and _mir_-124a, previously cloned from mouse (14); the others (not shown) with brain-restricted expression patterns include the rat homologue of _mir_-103 cloned from the Gemin3-Gemin4 - eIF2C2 complex in human HeLa cells (16), _mir_-128 from mouse (14), and four previously uncharacterized miRNAs (_mir_-323,-326, -329, and -344). Four miRNAs exhibit a broader expression pattern. Two miRNAs are shown in b: _mir_-191 and _mir_-_324_-5p; the others with broad expression patterns were _mir_-335 and _mir_-151*. The tissues analyzed were cortex (cx), cerebellum (cb), kidney (kd), heart (ht), liver (lv), thymus (th), lung (lu), ovary (ov), and testis (ts).
Fig. 3.
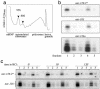
Localization of miRNAs to the mRNP and polyribosomes in cortical neurons. (a) Cytoplasmic extracts of primary cortical neurons were lysed and resolved on 15-45% sucrose gradients after sedimentation of nuclei and the majority of mitochondria. Continuous RNA measurements (_A_254) of the gradient fractions were measured. (b) Fraction 1 contains mRNPs, fractions 2 and 3 contain 55S and 80S ribosomal subunits, respectively, fraction 4 contains polyribosomes, and fraction 5 contains RNA granules. Total RNA isolated from each gradient fraction was analyzed by Northern blots with complementary probes for 8 miRNAs [_mir_-_129_-2*, -103, and -124a (shown) and _mir_-128,-323,-326,-329, and 344 (not shown)]. All of the miRNAs tested are predominantly localized to the mRNP and polyribosome fractions. (c) Four miRNAs [_mir_-128 and -_129_-2* (shown) and _mir_-326 and -344 (not shown)] were analyzed on cortical neuron cultures that were untreated (0′) or incubated in 50 mM KCl for 10 min (10′) or 2 h (120′) and resolved by sucrose density gradients as in b. A change in miRNA expression or distribution in the mRNP and polyribosome fractions was not detected after KCl treatment for these miRNAs.
Fig. 4.
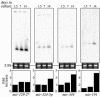
Temporal expression of miRNAs in primary rat cortical cultures. Dissociated neurons from E18 rat forebrain were plated and harvested after 1.5, 7, and 14 days in culture. Northern blots of equal amounts of total RNA isolated from these primary cultures were assayed with probes against 12 miRNAs. Three of seven miRNAs (_mir_-191, -_324_-5p, and -344) that increase in expression level from day 1.5 to day 14 cultures are shown (the remaining miRNAs, _mir_-128, -323, -326, and -329, are not shown). _mir_-_129_-2* (shown) is among five miRNAs tested (_mir_-103,-124a,-335,-_129_-2*, and -151*) that display a relatively constant level of expression. The quantitation of the miRNA expression levels was normalized to day 1.5 cultures.
Similar articles
- Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation.
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Sempere LF, et al. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. Epub 2004 Feb 16. Genome Biol. 2004. PMID: 15003116 Free PMC article. - miRNAs get an early start on translational silencing.
Meister G. Meister G. Cell. 2007 Oct 5;131(1):25-8. doi: 10.1016/j.cell.2007.09.021. Cell. 2007. PMID: 17923084 Review. - MicroRNAs and mammalian ovarian development.
Zhao H, Rajkovic A. Zhao H, et al. Semin Reprod Med. 2008 Nov;26(6):461-8. doi: 10.1055/s-0028-1096126. Epub 2008 Oct 24. Semin Reprod Med. 2008. PMID: 18951328 Review. - miRNP:mRNA association in polyribosomes in a human neuronal cell line.
Nelson PT, Hatzigeorgiou AG, Mourelatos Z. Nelson PT, et al. RNA. 2004 Mar;10(3):387-94. doi: 10.1261/rna.5181104. RNA. 2004. PMID: 14970384 Free PMC article. - MicroRNAs, mRNAs, and translation.
Maroney PA, Yu Y, Nilsen TW. Maroney PA, et al. Cold Spring Harb Symp Quant Biol. 2006;71:531-5. doi: 10.1101/sqb.2006.71.043. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381336 Review.
Cited by
- Control of mitochondrial activity by miRNAs.
Li P, Jiao J, Gao G, Prabhakar BS. Li P, et al. J Cell Biochem. 2012 Apr;113(4):1104-10. doi: 10.1002/jcb.24004. J Cell Biochem. 2012. PMID: 22135235 Free PMC article. Review. - Microarray analysis of microRNA expression in the developing mammalian brain.
Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Miska EA, et al. Genome Biol. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. Epub 2004 Aug 31. Genome Biol. 2004. PMID: 15345052 Free PMC article. - Genomics and the future of psychopharmacology: MicroRNAs offer novel therapeutics .
Gurwitz D. Gurwitz D. Dialogues Clin Neurosci. 2019;21(2):131-148. doi: 10.31887/DCNS.2019.21.2/dgurwitz. Dialogues Clin Neurosci. 2019. PMID: 31636487 Free PMC article. - microRNAs Sculpt Neuronal Communication in a Tight Balance That Is Lost in Neurological Disease.
Thomas KT, Gross C, Bassell GJ. Thomas KT, et al. Front Mol Neurosci. 2018 Dec 12;11:455. doi: 10.3389/fnmol.2018.00455. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30618607 Free PMC article. Review. - MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome.
Zhou Q, Souba WW, Croce CM, Verne GN. Zhou Q, et al. Gut. 2010 Jun;59(6):775-84. doi: 10.1136/gut.2009.181834. Epub 2009 Dec 1. Gut. 2010. PMID: 19951903 Free PMC article.
References
- Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. - PubMed
- Wightman, B., Ha, I. & Ruvkun, G. (1993) Cell 75, 855-862. - PubMed
- Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. & Ruvkun, G. (2000) Nature 403, 901-906. - PubMed
- Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R. & Ruvkun, G. (2000) Mol. Cell 5, 659-669. - PubMed
- Rougvie, A. E. (2001) Nat. Rev. Genet. 2, 690-701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources