Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping - PubMed (original) (raw)
Comparative Study
Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping
Martha S Rook et al. Am J Pathol. 2004 Jan.
Abstract
Genome-wide screening of genetic alterations between normal and cancer cells, as well as among subgroups of tumors, is important for establishing molecular mechanism and classification of cancer. Gene silencing through loss of heterozygosity is widely observed in cancer cells and detectable by analyzing allelic loss of single nucleotide polymorphism and/or short tandem repeat markers. To use minute quantities of DNA that are available through laser capture microdissection (LCM) of cancer cells, a whole genome amplification method that maintains locus and allele balance is essential. We have successfully used a ø29 polymerase-based isothermal whole genome amplification method to amplify LCM DNA using a proteinase K lysis procedure coupled with a pooling strategy. Through single nucleotide polymorphism and short tandem repeat genotype analysis we demonstrate that using pooled DNA from two or three separate amplification reactions significantly reduces any allele bias introduced during amplification. This strategy is especially effective when using small quantities of source DNA. Although a convenient alkaline lysis DNA extraction procedure provided satisfactory results from using 1500 to 3000 LCM cells, proteinase K digestion was superior for lower cell numbers. Accurate genotyping is achieved with as few as 100 cells when both proteinase K extraction and pooling are applied.
Figures
Figure 1
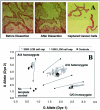
A: Representative LCM images of a colon cancer cell microdissection. B: TaqMan SNP-genotyping assay plot for G/A1182 of the EDNRB gene with WGA products from 70 different 3000 prostate LCM cell caps and WGA products from 12 different 1500 colon LCM cell caps. Assays were done in replicates. Arbitrary fluorescence units are shown on axes.
Figure 2
Schematic sample preparation and data analysis.
Figure 3
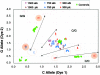
Effect of lysis procedure on a TaqMan SNP-genotyping assay for C/G16996 of the ERBB3 gene. LCM colon cell caps containing 1500 cells (8 caps), 750 cells (12 caps), and 300 cells (8 caps) were lysed by a proteinase K (pk) or an alkaline (alk) lysis protocol. All samples should give a heterozygote call. No calls are highlighted by open circles and miscalls are highlighted by filled circles. Shown on axes are arbitrary fluorescence units.
Figure 4
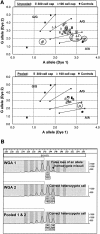
A: Effect of pooling WGA products from a 300 or a 100 LCM prostate cell cap on TaqMan SNP-genotyping assay for A/G36177 of the CYP3A5 gene: 24 WGA products were assayed in replicates for each cell number to afford 96 unpooled data points; 8 individual WGA products were assayed in replicates for each cell number to afford 32 pooled data points. No calls are highlighted by open circles and miscalls are highlighted by filled circles. Shown on axes are arbitrary fluorescence units. B: Effect of pooling WGA products from a 300 colon cell cap on a STR-genotyping assay using a 2-bp repeat marker D13S173.
Figure 5
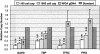
Relative gene copy number analysis using an unamplified genomic DNA as a standard. Pooled WGA products from microdissected colon cells were analyzed using TaqMan quantitative PCR assays and compared with a WGA product from 5 ng of genomic DNA isolated from cultured cells. The copy numbers are averages of four or six amplified samples, and the error bars show corresponding SD.
Similar articles
- Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.
Shabaninejad H, Kenny RP, Robinson T, Stoniute A, O'Keefe H, Still M, Thornton C, Pearson F, Beyer F, Meader N. Shabaninejad H, et al. Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article. - Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.
Pillay J, Gaudet LA, Saba S, Vandermeer B, Ashiq AR, Wingert A, Hartling L. Pillay J, et al. Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Pharmacological treatments in panic disorder in adults: a network meta-analysis.
Guaiana G, Meader N, Barbui C, Davies SJ, Furukawa TA, Imai H, Dias S, Caldwell DM, Koesters M, Tajika A, Bighelli I, Pompoli A, Cipriani A, Dawson S, Robertson L. Guaiana G, et al. Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
- Performance of whole-genome amplified DNA isolated from serum and plasma on high-density single nucleotide polymorphism arrays.
Croft DT Jr, Jordan RM, Patney HL, Shriver CD, Vernalis MN, Orchard TJ, Ellsworth DL. Croft DT Jr, et al. J Mol Diagn. 2008 May;10(3):249-57. doi: 10.2353/jmoldx.2008.070155. Epub 2008 Apr 10. J Mol Diagn. 2008. PMID: 18403606 Free PMC article. - Amplification of whole tumor genomes and gene-by-gene mapping of genomic aberrations from limited sources of fresh-frozen and paraffin-embedded DNA.
Bredel M, Bredel C, Juric D, Kim Y, Vogel H, Harsh GR, Recht LD, Pollack JR, Sikic BI. Bredel M, et al. J Mol Diagn. 2005 May;7(2):171-82. doi: 10.1016/S1525-1578(10)60543-0. J Mol Diagn. 2005. PMID: 15858140 Free PMC article. - Whole genome amplification for array comparative genomic hybridization using DNA extracted from formalin-fixed, paraffin-embedded histological sections.
Huang J, Pang J, Watanabe T, Ng HK, Ohgaki H. Huang J, et al. J Mol Diagn. 2009 Mar;11(2):109-16. doi: 10.2353/jmoldx.2009.080143. Epub 2009 Feb 5. J Mol Diagn. 2009. PMID: 19197000 Free PMC article. - Simultaneous isolation of DNA and RNA from the same cell population obtained by laser capture microdissection for genome and transcriptome profiling.
Xu C, Houck JR, Fan W, Wang P, Chen Y, Upton M, Futran ND, Schwartz SM, Zhao LP, Chen C, Mendez E. Xu C, et al. J Mol Diagn. 2008 Mar;10(2):129-34. doi: 10.2353/jmoldx.2008.070131. Epub 2008 Feb 7. J Mol Diagn. 2008. PMID: 18258925 Free PMC article. - Whole genome amplification and its impact on CGH array profiles.
Talseth-Palmer BA, Bowden NA, Hill A, Meldrum C, Scott RJ. Talseth-Palmer BA, et al. BMC Res Notes. 2008 Jul 29;1:56. doi: 10.1186/1756-0500-1-56. BMC Res Notes. 2008. PMID: 18710509 Free PMC article.
References
- Weber BL. Cancer genomics. Cancer Cell. 2002;1:37–47. - PubMed
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. Lymphoma/Leukemia Molecular Profiling Project: The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. - PubMed
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. - PubMed
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. - PubMed
- Thiagalingam S, Foy RL, Cheng KH, Lee HJ, Thiagalingam A, Ponte JF. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Curr Opin Oncol. 2002;14:65–72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources