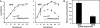A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms - PubMed (original) (raw)
A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms
Damien Portevin et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Mycolic acids are major and specific constituents of the cell envelope of Corynebacterineae, a suborder of bacterial species including several important human pathogens such as Mycobacterium tuberculosis, Mycobacterium leprae, or Corynebacterium diphtheriae. These long-chain fatty acids are involved in the unusual architecture and impermeability of the cell envelope of these bacteria. The condensase, the enzyme responsible for the final condensation step in mycolic acid biosynthesis, has remained an enigma for decades. By in silico analysis of various mycobacterial genomes, we identified a candidate enzyme, Pks13, that contains the four catalytic domains required for the condensation reaction. Orthologs of this enzyme were found in other Corynebacterineae species. A Corynebacterium glutamicum strain with a deletion in the pks13 gene was shown to be deficient in mycolic acid production whereas it was able to produce the fatty acids precursors. This mutant strain displayed an altered cell envelope structure. We showed that the pks13 gene was essential for the survival of Mycobacterium smegmatis. A conditional M. smegmatis mutant carrying its only copy of pks13 on a thermosensitive plasmid exhibited mycolic acid biosynthesis defect if grown at nonpermissive temperature. These results indicate that Pks13 is the condensase, a promising target for the development of new antimicrobial drugs against Corynebacterineae.
Figures
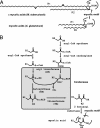
Fig. 1.
Structure of mycolic acids and proposed terminal steps in their biosynthesis. (A) Structure of mycolic acids from M. tuberculosis (α type) and C. glutamicum. These 2-alkyl 3-hydroxyl fatty acids are elaborated by members of the Corynebacterineae suborder; their chain lengths vary from C30 to C90 according to the bacterial genera. In mycobacteria, cis and/or trans cyclopropyl groups and double bonds may be found on the main chain (R1), leading to the so-called α-mycolates; two additional types of mycolates, with a keto and methoxy groups located on R1, are also produced by M. tuberculosis. (B) Proposed pathway for the condensation of two fatty acids to form mycolic acids (9). R1 and R2 chains vary according to the Corynebacterineae species (see A).
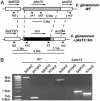
Fig. 2.
Construction of a C. glutamicum Δpks13::km mutant. (A) Schematic representation of the genetic structure of the pks13 locus in WT C. glutamicum and in the Δpks13::km mutant. The boxes indicate the various genes of the pks13 locus. The binding sites (arrow head) and names of the primers used for the PCR analysis of the mutant strains are indicated. The expected PCR amplification products for the various strains are indicated below each genetic structure. (B) PCR analysis showed that the C. glutamicum Δpks13::km mutant contains a km cassette within pks13.
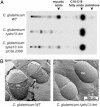
Fig. 3.
Biochemical and ultrastructural analysis of the envelope of C. glutamicum Δpks13::km mutant. (A) Fatty acid contents of WT C. glutamicum, the Δpks13::km mutant strain, and the Δpks13::km mutant strain, complemented with a plasmid carrying pks13, were evaluated on TLC after labeling with [14C]acetate. (B) Electron microscopy observation of freeze-fractured bacterial cultures of WT C. glutamicum or the Δpks13::km mutant. A fracture plan was observed in the cell wall (CW) for the WT strain and in the plasma membrane (PM) for the Δpks13::km mutant.
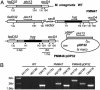
Fig. 4.
Construction of a conditional mutant of M. smegmatis.(A) Schematic representation of the genetic structure of the pks13 genomic locus obtained during construction of the conditional M. smegmatis mutant. The boxes indicate the various genes of the pks13 locus. The binding sites (arrowheads) and names of the primers used for PCR analysis of the mutant strains are indicated. The expected PCR amplification products for the various strains are indicated below the last genetic structure. (B) PCR analysis of the conditional mutant PMM48:pDP32 and its parental strains PMM47 and mc2155.
Fig. 5.
Growth characteristics and mycolic acid contents of the conditional M. smegmatis mutant incubated at 30°C and 42°C. (A) Colony-forming units (cfu) counted during growth of the WT strain or PMM48:pDP32 at 30°C or 42°C. The values shown are the means ± SD for three independant experiments. (B) Comparison of mycolates contents of WT and the conditional Δpks13::hyg mutant after growth at permissive temperature (30°C) or shift to nonpermissive temperature (42°C). The mycolate:C16-C18 fatty acids ratio was quantified for the WT strain and PMM48:pDP32. The ratio obtained for PMM48:pDP32 was divided by that obtained for the WT strain in the same growth conditions. The values shown are the means ± SD for three independant determinations.
Similar articles
- The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein-protein interactions.
Veyron-Churlet R, Bigot S, Guerrini O, Verdoux S, Malaga W, Daffé M, Zerbib D. Veyron-Churlet R, et al. J Mol Biol. 2005 Nov 4;353(4):847-58. doi: 10.1016/j.jmb.2005.09.016. Epub 2005 Sep 23. J Mol Biol. 2005. PMID: 16213523 - The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis.
Sacco E, Covarrubias AS, O'Hare HM, Carroll P, Eynard N, Jones TA, Parish T, Daffé M, Bäckbro K, Quémard A. Sacco E, et al. Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14628-33. doi: 10.1073/pnas.0704132104. Epub 2007 Sep 5. Proc Natl Acad Sci U S A. 2007. PMID: 17804795 Free PMC article. - Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis.
Takayama K, Wang C, Besra GS. Takayama K, et al. Clin Microbiol Rev. 2005 Jan;18(1):81-101. doi: 10.1128/CMR.18.1.81-101.2005. Clin Microbiol Rev. 2005. PMID: 15653820 Free PMC article. Review. - [Mycolic acids--potential biomarkers of opportunistic infections caused by bacteria of the suborder Corynebacterineae].
Kowalski K, Szewczyk R, Druszczyńska M. Kowalski K, et al. Postepy Hig Med Dosw (Online). 2012 Jun 29;66:461-8. doi: 10.5604/17322693.1002082. Postepy Hig Med Dosw (Online). 2012. PMID: 22922146 Review. Polish.
Cited by
- Identification of valine- or leucine-containing glycopeptidolipids from Mycobacterium avium-intracellulare complex.
Ichimura N, Kasama T. Ichimura N, et al. Curr Microbiol. 2012 Jun;64(6):561-8. doi: 10.1007/s00284-012-0107-6. Epub 2012 Mar 22. Curr Microbiol. 2012. PMID: 22437852 - Clinical strains of Mycobacterium tuberculosis exhibit differential lipid metabolism-associated transcriptome changes in in vitro cholesterol and infection models.
Moopanar K, Nyide ANG, Senzani S, Mvubu NE. Moopanar K, et al. Pathog Dis. 2023 Jan 17;81:ftac046. doi: 10.1093/femspd/ftac046. Pathog Dis. 2023. PMID: 36509392 Free PMC article. - Proton transfer activity of the reconstituted Mycobacterium tuberculosis MmpL3 is modulated by substrate mimics and inhibitors.
Stevens CM, Babii SO, Pandya AN, Li W, Li Y, Mehla J, Scott R, Hegde P, Prathipati PK, Acharya A, Liu J, Gumbart JC, North J, Jackson M, Zgurskaya HI. Stevens CM, et al. Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2113963119. doi: 10.1073/pnas.2113963119. Epub 2022 Jul 19. Proc Natl Acad Sci U S A. 2022. PMID: 35858440 Free PMC article. - Novel Polyoxyethylene-Containing Glycolipids Are Synthesized in Corynebacterium matruchotii and Mycobacterium smegmatis Cultured in the Presence of Tween 80.
Wang C, Mahrous EA, Lee RE, Vestling MM, Takayama K. Wang C, et al. J Lipids. 2011;2011:676535. doi: 10.1155/2011/676535. Epub 2010 Jul 20. J Lipids. 2011. PMID: 21490808 Free PMC article. - Bacterial Cell Wall Modification with a Glycolipid Substrate.
Calabretta PJ, Hodges HL, Kraft MB, Marando VM, Kiessling LL. Calabretta PJ, et al. J Am Chem Soc. 2019 Jun 12;141(23):9262-9272. doi: 10.1021/jacs.9b02290. Epub 2019 Jun 4. J Am Chem Soc. 2019. PMID: 31081628 Free PMC article.
References
- Anonymous (2000) The WHO/IUATLD Global Project on Antituberculosis Drug Resistance Surveillance (World Health Organization, Geneva).
- Webb, V. & Davies, J. (1999) in Mycobacteria: Molecular Biology and Virulence, eds. Ratledge, C. & Dale, J. (Blackwell Science, Oxford, U.K.), Vol. 1, pp. 287-307.
- Daffé, M. & Draper, P. (1998) Adv. Microbiol. Physiol. 39, 131-203. - PubMed
- Brennan, P. J. & Nikaido, H. (1995) Annu. Rev. Biochem. 64, 29-63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous