Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression - PubMed (original) (raw)
Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression
Norihisa Ooe et al. Mol Cell Biol. 2004 Jan.
Abstract
Sim2, a basic helix-loop-helix (bHLH)-PAS transcriptional repressor, is thought to be involved in some symptoms of Down's syndrome. In the course of searching for hypothetical Sim2 relatives, we isolated another bHLH-PAS factor, NXF. NXF was a novel gene and was selectively expressed in neuronal tissues. While no striking homolog of NXF was found in vertebrates, a Caenorhabditis elegans putative transcription factor, C15C8.2, showed similarity in the bHLH-PAS domain. NXF had an activation domain as a transcription activator, and Arnt-type bHLH-PAS subfamily members were identified as the heterodimer partners of NXF. The NXF/Arnt heterodimer was capable of binding and activating a subset of Sim2/Arnt target DNA variants, and Sim2 could compete with the NXF activity on the elements. We showed that Drebrin had several such NXF/Arnt binding elements on the promoter, which could be direct or indirect cross talking points between NXF (activation) and Sim2 (repression) action. Drebrin has been reported to be engaged in dendritic-cytoskeleton modulation at synapses, and such a novel NXF signaling system on neural gene promoter may be a molecular target of the adverse effects of Sim2 in the mental retardation of Down's syndrome.
Figures
FIG. 1.
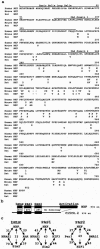
NXF has a bHLH-PAS domain. (a) Predicted amino acid sequences determined from human, mouse, and rat NXF cDNAs. Sequence deletions are indicated by dashes. For the mouse and rat NXF sequences, only differences are shown. (b) Comparison of human NXF with the C. elegans C15C8.2 (putative bHLH-PAS transcription-factor; accession no. AF370361), demonstrating the greatest homology. The percent amino acid identity is indicated for the bHLH, PAS1, and PAS2 domains. There is no homology in the C terminus. (c) Percent amino acid identity between NXF and mammalian bHLH-PAS factors in each domain. Only the three factors most similar to NXF in homology searches are shown for each domain.
FIG. 2.
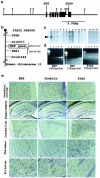
Chromosomal location and expression pattern of NXF. (a) Structural organization of the mouse NXF gene. The gene consists of eight exons shown (filled boxes) covering about 5 kbp of the genome. H, _Hin_dIII; X, _Xho_I. (b) NXF gene loci in a computerized database of the human chromosome STS map maintained by the National Center for Biotechnology Information (
). (c) RNA blotting analysis of NXF expression in human adult tissues. Lanes: 1, brain; 2, heart; 3, skeletal muscle; 4, colon; 5, thymus; 6, spleen; 7, kidney; 8, liver; 9, small intestine; 10, placenta; 11, lung; 12, leukocyte; 13, prostate; 14, testis; 15, uterus. (d) RT-PCR analysis of NXF and Sim2 mRNAs in developing mouse embryos. Each target was amplified by PCR with an equal amount of mRNA from a 7-day embryo (lane 1), an 11-day embryo (lane 2), a 15-day embryo (lane 3), a 17-day embryo (lane 4), and 4-week postnatal brain (lane 5). (e) In situ hybridization analysis of NXF, Drebrin, and Sim2 expression in several brain regions (from an 8-week-old rat). The sections were hybridized with antisense RNA probes from the respective gene cDNAs. Violet signals indicate the dotted pattern of expression of each gene. The sense probe detected no signals.
FIG. 3.
Properties of NXF as a transcriptional regulator. (a) Gal4-responsive reporter gene (Gal-Rex4) activation in mammalian cells with NXF C-terminal fragment fused Gal4 DNA binding domain effectors. Gal4-DBD expresses the Gal4 DNA binding domain without the activation domain. Gal-NXF(C)expresses the fusion protein between this Gal4 DNA binding domain and the NXF C terminus (amino acids 256 to 802). Likewise, Gal-NXF(256-596) and Gal-NXF(597-802) express Gal4-DBD fusion proteins with NXF amino acids 256 to 596 and 597 to 802, respectively. (b) NXF interacts with other bHLH-PAS factors as a heterodimer, as assessed by mammalian two-hybrid analysis. Panel 1, the Gal-NXF(N) bait is the Gal4-DBD fusion protein with NXF amino acids 1 to 596 containing the N-terminal bHLH-PAS domain. Each VP16-X is the VP16 activation domain fusion protein with the bHLH-PAS domain from the respective factors. CP, control protein (viral coat protein) unrelated to bHLH-PAS. Data are means and standard deviations of fold induction relative to the control (bar 1) (n = 4). Panels 2 and 3, The control bait protein Gal-Sim2(N) or Gal-Clock(N) bait is a Gal4-DBD fusion protein with the Sim2 N-terminal bHLH-PAS domain or Clock N-terminal bHLH-PAS domain, respectively. These baits confirmed the known interactions.
FIG. 4.
DNA binding selectivity of NXF and potential cross talking with Sim2. (a) DNA binding activity with CME variants which are mutated one by one at the first position and the second position next to the boxed GTG in the CME core and with additional variants containing ACGTG (designated CME), GCGTG, TCGTG, or CCGTG as the core sequence. The boxed GTG in the CME core element is known as an Arnt binding sequence. Combinations of recombinant baculovirus expressing Arnt2 and NXF, along with control lysate (wild-type virus infected), were used. (b) Confirmation of potential CME binding activity of NXF/Arnt2. The typical Sim2-responsive element CME probe also shows significant binding activity with NXF/Arnt2 protein. +Cold CME, excess unlabeled probe added in the competition experiment. The E-box is excess unlabeled probe distinct from CME. (c) Confirmation of NXF- and Arnt2-dependent transcriptional activation of the CME regulatory element in reporter analysis. Data are means (n = 4) and standard deviations. Arrowheads, NXF/Arnt2 binding activity; asterisk, Arnt2 homodimer binding activity. (d) Comparison of transcriptional regulatory activity of NXF and Sim2 oneach reporter with the mutated core in the parental CME element unit on the promoter. In the Sim2-VP16 effector, the Sim2 transcriptional repression domain was replaced with the VP16 activation domain, and it could act as transcriptional activator showing Sim2 DNA binding specificity. Data are means (n = 4) and standard deviations.
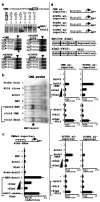
FIG. 5.
Identification of NXF target genes. (a) The Drebrin gene and the NXF gene itself are induced in the NXF-overexpressing neuroblastoma SK-N-MC, while the negative control Sim2 gene promoter has constant mRNA expression with or without NXF. (b) The NXF and Drebrin gene promoters are activated by NXF/Arnt2 and repressed by Sim2/Arnt2 in reporter analysis (data are means [n = 4] and standard deviations of fold induction, relative to cells transfected with the reporter gene alone). (c) NXF and Drebrin gene 5′ flanking regions. Putative NXF responsive elements are numbered. The filled box is exon 1. (d) NXF/Arnt2 binds directly to some of the listed candidate elements from the NXF or Drebrin promoter and activates some reporter gene constructs having two copies of each element (20-mer). The numbered oligonucleotide sequences are derived from the NXF or Drebrin promoter, and the numbering of each element is the same as in panel c. The fold induction values for NXF/Arnt2 activities relative to that with Arnt2 alone are listed on the right (means and standard deviations [s.d.]; n = 4). Arrowhead, specific binding signal with each labeled oligonucleotide; asterisk, Arnt2 homodimer; #, unreliable values due to high background.
FIG. 6.
Confirmation of the NXF and Drebrin promoters as direct targets of the NXF/Arnt2 complex. (a) Mutational analysis of putative NXF/Arnt2 binding elements on the NXF or Drebrin promoter. The mutated NXF promoter here was point mutated (from GTG in each core to AAG) only at strong NXF/Arnt2 binding sites (three locations) among putative elements. The numbering of putative binding sites is the same as in Fig. 5c. Mutated Drebrin promoter A (Mutated A) has point mutations only at the elements (three locations) which did not show an NXF/Arnt2 binding signal in our DNA binding assay (Fig. 5d). Mutated Drebrin promoter B is mutated only at the elements which show a significant NXF/Arnt2 binding signal in Fig. 5d. Each promoter activity was analyzed in a reporter assay with or without NXF/Arnt2 complex. (b) NXF directly binds to the Drebrin promoter in vivo. The endogenous Drebrin promoter chromatin fragment was specifically immunoprecipitated by Flag-NXF in a sonicated chromatin source of Flag-NXF/Arnt2-overexpressing transformants. The immunoprecipitated chromatin fragment was detected by PCR. PCR primer A is for the Drebrin promoter region, and PCR primer B, which amplifies the Arnt2 exon, is used for a negative control. Input for the positive control is the sonicated chromatin source before anti-Flag immunoprecipitation.
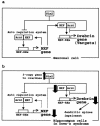
FIG. 7.
(a) Proposed model of the action of NXF in neural target gene transcriptional regulation. (b) Proposed hypothesis for a molecular basis of the action of NXF and Sim2 in nerve cells such as hippocampal neurons, related to mental retardation with DS.
Similar articles
- Characterization of Drosophila and Caenorhabditis elegans NXF-like-factors, putative homologs of mammalian NXF.
Ooe N, Saito K, Oeda K, Nakatuka I, Kaneko H. Ooe N, et al. Gene. 2007 Oct 1;400(1-2):122-30. doi: 10.1016/j.gene.2007.06.005. Epub 2007 Jun 14. Gene. 2007. PMID: 17628356 - cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt).
Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. Hirose K, et al. Mol Cell Biol. 1996 Apr;16(4):1706-13. doi: 10.1128/MCB.16.4.1706. Mol Cell Biol. 1996. PMID: 8657146 Free PMC article. - Aryl hydrocarbon receptor-mediated signal transduction.
Rowlands JC, Gustafsson JA. Rowlands JC, et al. Crit Rev Toxicol. 1997 Mar;27(2):109-34. doi: 10.3109/10408449709021615. Crit Rev Toxicol. 1997. PMID: 9099515 Review. - Structural and functional analysis of hypoxia-inducible factor 1.
Semenza GL, Agani F, Booth G, Forsythe J, Iyer N, Jiang BH, Leung S, Roe R, Wiener C, Yu A. Semenza GL, et al. Kidney Int. 1997 Feb;51(2):553-5. doi: 10.1038/ki.1997.77. Kidney Int. 1997. PMID: 9027737 Review.
Cited by
- Npas4 Transcription Factor Expression Is Regulated by Calcium Signaling Pathways and Prevents Tacrolimus-induced Cytotoxicity in Pancreatic Beta Cells.
Speckmann T, Sabatini PV, Nian C, Smith RG, Lynn FC. Speckmann T, et al. J Biol Chem. 2016 Feb 5;291(6):2682-95. doi: 10.1074/jbc.M115.704098. Epub 2015 Dec 9. J Biol Chem. 2016. PMID: 26663079 Free PMC article. - NPAS4 Exacerbates Pyroptosis via Transcriptionally Regulating NLRP6 in the Acute Phase of Intracerebral Hemorrhage in Mice.
Jian D, Qin L, Gan H, Zheng S, Xiao H, Duan Y, Zhang M, Liang P, Zhao J, Zhai X. Jian D, et al. Int J Mol Sci. 2023 May 5;24(9):8320. doi: 10.3390/ijms24098320. Int J Mol Sci. 2023. PMID: 37176030 Free PMC article. - Upregulation of Npas4 protein expression by chronic administration of amphetamine in rat nucleus accumbens in vivo.
Guo ML, Xue B, Jin DZ, Liu ZG, Fibuch EE, Mao LM, Wang JQ. Guo ML, et al. Neurosci Lett. 2012 Oct 24;528(2):210-4. doi: 10.1016/j.neulet.2012.07.048. Epub 2012 Aug 3. Neurosci Lett. 2012. PMID: 22884934 Free PMC article. - Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity.
Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Zhang SJ, et al. PLoS Genet. 2009 Aug;5(8):e1000604. doi: 10.1371/journal.pgen.1000604. Epub 2009 Aug 14. PLoS Genet. 2009. PMID: 19680447 Free PMC article. - Human variants in the neuronal basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor complex NPAS4/ARNT2 disrupt function.
Bersten DC, Bruning JB, Peet DJ, Whitelaw ML. Bersten DC, et al. PLoS One. 2014 Jan 17;9(1):e85768. doi: 10.1371/journal.pone.0085768. eCollection 2014. PLoS One. 2014. PMID: 24465693 Free PMC article.
References
- Becker, L. E., D. L. Armstrong, and F. Chan. 1986. Dendritic atrophy in children with Down's syndrome. Ann. Neurol. 20:520-526. - PubMed
- Becker, L., T. Mito, S. Takashima, and K. Onodera. 1991. Growth and development of the brain in Down syndrome. Prog. Clin. Biol. Res. 373:133-152. - PubMed
- Brunskill, E. W., D. P. Witte, A. B. Shreiner, and S. S. Potter. 1999. Characterization of Npas3, a novel basic helix-loop-helix gene expressed in the developing mouse nervous system. Mech. Dev. 88:237-241. - PubMed
- Chen, H., R. Chrast, C. Rossier, A. Gos, S. E. Antonarakis, J. Kudoh, N. Shindoh, H. Maeda, S. Minoshima, and N. Shimizu. 1995. Single-minded and Down syndrome? Nat. Genet. 10:9-10. - PubMed
- Chrast, R., H. S. Scott, R. Madani, L. Huber, D. P. Wolfer, M. Prinz, A. Aguzzi, H.-P. Lipp, and S. E. Antonarakis. 2000. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9:1853-1864. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases