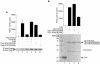Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A - PubMed (original) (raw)
Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A
Heehyoung Lee et al. Mol Cell Biol. 2004 Jan.
Abstract
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl groups from lysine residues of histone and nonhistone proteins. Recent studies suggest that they are key regulators of many cellular events, including cell proliferation and cancer development. Human class I HDACs possess homology to the yeast RPD3 protein and include HDAC1, HDAC2, HDAC3, and HDAC8. While HDAC1, HDAC2, and HDAC3 have been characterized extensively, almost nothing is known about HDAC8. Here we report that HDAC8 is phosphorylated by cyclic AMP-dependent protein kinase A (PKA) in vitro and in vivo. The PKA phosphoacceptor site of HDAC8 is Ser(39), a nonconserved residue among class I HDACs. Mutation of Ser(39) to Ala enhances the deacetylase activity of HDAC8. In contrast, mutation of Ser(39) to Glu or induction of HDAC8 phosphorylation by forskolin, a potent activator of adenyl cyclase, decreases HDAC8's enzymatic activity. Remarkably, inhibition of HDAC8 activity by hyperphosphorylation leads to hyperacetylation of histones H3 and H4, suggesting that PKA-mediated phosphorylation of HDAC8 plays a central role in the overall acetylation status of histones.
Figures
FIG. 1.
Amino acid sequences of HDAC8. (A) Amino acid sequence of human HDAC8 (AF230097). Residues within the putative catalytic domain are highlighted in bold. Potential phosphorylation sites, as determined by phosphobase detection (
http://www.cbs.dtu.dk/databases/PhosphoBase/predict/predict.html
), are underlined. The potential PKA phosphorylation site (Ser39) is marked with an asterisk. (B) Top, a potential PKA phosphorylation site in HDAC8 determined by database sequence analysis. Middle and bottom, comparison of the human HDAC8 sequence surrounding Ser39 with other human class I HDAC sequences and with the mouse HDAC8 sequence.
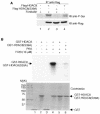
FIG. 2.
In vivo phosphorylation of HDAC8. HeLa cells were infected with a recombinant adenovirus that expresses Flag-HDAC8 and were labeled with 32Pi for 4 h. Cell extracts were immunoprecipitated with an anti-Flag antibody, and immunoprecipitates (IP) were resolved on an SDS-8% polyacrylamide gel. Phosphoproteins were visualized by autoradiography. As a negative control (lane 1), cells were infected with an adenovirus that expresses GFP alone. In lanes 3 and 4, cells were treated with the indicated dose of H-89 for 45 min before harvest. The positions of the molecular weight markers (weights are in thousands) are indicated on the left. Immunoprecipitates were immunoblotted with an anti-Flag antibody to ensure equal amounts of Flag-HDAC8 in lanes 2 to 4 (bottom panel). IgH, immunoglobulin H.
FIG. 3.
Stimulation of HDAC8 phosphorylation by forskolin. (A) With filter-binding assays, PKA activities were determined in extracts prepared from HeLa cells treated with 10 μM forskolin for 45 min in the presence or absence of H-89 (10 μM). (B) Bacterially expressed, purified GST-HDAC8 was incubated with HeLa cell extracts prepared from cells that were untreated or treated with 10 μM forskolin for 45 min. H-89 (10 μM) was added to the in vitro phosphorylation reaction mixtures as indicated (lane 4). GST was used as a negative control (lane 5).
FIG. 4.
In vitro phosphorylation of HDAC8 with purified PKA. (A) Purified GST-HDAC8 was incubated with PKA in the presence or absence of H-89 (0.15 μM in lanes 4 and 9, 2 μM in lanes 5 and 10, 10 μM in lanes 6 and 11), and in vitro kinase reactions were performed. Proteins were resolved by SDS-PAGE, and 32P-labeled proteins were visualized by autoradiography. GST (lane 1) and GST-H2B (lanes 2 to 6) were used as negative and positive controls, respectively. (B) GST-HDAC8 was phosphorylated by PKA in the presence of [γ-32P]ATP. Radiolabeled GST-HDAC8 was eluted from an SDS-polyacrylamide gel, digested with trypsin, and analyzed by TLC.
FIG. 5.
Identification of serine as the phosphoacceptor residue in HDAC8. (A) HDAC8 was phosphorylated by PKA in the presence of [γ-32P]ATP and eluted from an SDS-polyacrylamide gel. After partial acid hydrolysis, the sample was analyzed by cellulose TLC. P-Ser, P-Thr, and P-Tyr indicate the positions of phosphoserine, phosphothreonine, and phosphotyrosine, respectively. (B) HeLa cells were transfected with a plasmid encoding Flag-HDAC8 and treated with forskolin (10 μM) or H-89 (10 μM) as indicated. Cell extracts were immunoprecipitated (IP) with an anti-Flag antibody and immunoblotted (IB) with an anti-phosphoserine antibody (top) or an anti-Flag antibody (bottom). (C) Extracts prepared from HeLa cells treated with forskolin (10 μM) or PKI (10 μM) were immunoprecipitated with an anti-HDAC8 antibody or preimmune serum and immunoblotted with an anti-phosphoserine antibody (top) or an anti-HDAC8 antibody (bottom).
FIG. 6.
Identification of Ser39 as the phosphorylation site within HDAC8. (A) HeLa cells expressing Flag-HDAC8 or the Flag-HDAC8(S39A) mutant form were treated with forskolin (10 μM) as indicated. Cell extracts were immunoprecipitated (IP) with an anti-Flag antibody, and immune complexes were then immunoblotted (IB) with either an anti-phosphoserine antibody (top) or an anti-Flag antibody (bottom). (B) Purified GST-HDAC8 and GST-HDAC8(S39A) proteins were used as substrates for in vitro phosphorylation by PKA. The proteins were resolved by SDS-PAGE, and the 32P-radiolabeled proteins were visualized by autoradiography (top). Coomassie blue staining was performed before autoradiography to visualize the locations and amounts of the different proteins (bottom). The values on the left are molecular weights in thousands.
FIG. 7.
HDAC8 phosphorylation inhibits enzymatic activity. (A) HeLa cells were transfected with constructs expressing Flag-tagged wild-type or mutant HDAC8, as indicated. After treatment of cells with forskolin (10 μM) and H-89 (10 μM), Flag immunoprecipitates (IP) were assayed for HDAC activity. All experiments were normalized to equal amounts of DNA with parental expression vectors. Data shown are the average results ± the standard deviation from three separate transfections. Immunoprecipitates were immunoblotted (IB) with an anti-Flag antibody to ensure approximately equal amounts of Flag fusion proteins in reactions 2 to 6 (a representative blot is shown at the bottom). (B) Purified, GST, GST-HDAC8, GST-HDAC8(S39A), and GST-HDAC8(S39E) were assayed for deacetylase activity. All experiments were performed in triplicate, and the data shown are the average ± the standard deviation. Coomassie blue-stained gels were used to show approximately equal amounts of GST and GST fusion proteins used in each reaction mixture (a representative gel is shown at the bottom). The values on the left are molecular weights in thousands.
FIG. 8.
HDAC8 phosphorylation increases histone H3 and H4 acetylation. HeLa cells were transfected with plasmids expressing Flag-HDAC8 or Flag-HDAC8(S39A). After treatment with or without forskolin (10 μM), core histones were prepared and analyzed by a Western blot (immunoblot [IB]) assay with an anti-acetylated-H4 antibody (top). Subsequently, the blot was stripped and reprobed with an anti-acetylated-H3 antibody (middle). A separate SDS-polyacrylamide gel was prepared in parallel to assess the qualities of core histones in each reaction (bottom).
Similar articles
- HDAC8 substrates: Histones and beyond.
Wolfson NA, Pitcairn CA, Fierke CA. Wolfson NA, et al. Biopolymers. 2013 Feb;99(2):112-26. doi: 10.1002/bip.22135. Biopolymers. 2013. PMID: 23175386 Free PMC article. Review. - Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation.
Lee H, Sengupta N, Villagra A, Rezai-Zadeh N, Seto E. Lee H, et al. Mol Cell Biol. 2006 Jul;26(14):5259-69. doi: 10.1128/MCB.01971-05. Mol Cell Biol. 2006. PMID: 16809764 Free PMC article. - Cloning and characterization of a novel human histone deacetylase, HDAC8.
Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Buggy JJ, et al. Biochem J. 2000 Aug 15;350 Pt 1(Pt 1):199-205. Biochem J. 2000. PMID: 10926844 Free PMC article. - Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor.
Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, Johanson K, Sung CM, Liu R, Winkler J. Hu E, et al. J Biol Chem. 2000 May 19;275(20):15254-64. doi: 10.1074/jbc.M908988199. J Biol Chem. 2000. PMID: 10748112 - Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review.
Cited by
- HDAC8 substrates: Histones and beyond.
Wolfson NA, Pitcairn CA, Fierke CA. Wolfson NA, et al. Biopolymers. 2013 Feb;99(2):112-26. doi: 10.1002/bip.22135. Biopolymers. 2013. PMID: 23175386 Free PMC article. Review. - Lysine deacetylase inhibition promotes relaxation of arterial tone and C-terminal acetylation of HSPB6 (Hsp20) in vascular smooth muscle cells.
Chen A, Karolczak-Bayatti M, Sweeney M, Treumann A, Morrissey K, Ulrich SM, Europe-Finner GN, Taggart MJ. Chen A, et al. Physiol Rep. 2013 Nov;1(6):e00127. doi: 10.1002/phy2.127. Epub 2013 Nov 7. Physiol Rep. 2013. PMID: 24400135 Free PMC article. - TLR2-dependent inhibition of macrophage responses to IFN-gamma is mediated by distinct, gene-specific mechanisms.
Benson SA, Ernst JD. Benson SA, et al. PLoS One. 2009 Jul 24;4(7):e6329. doi: 10.1371/journal.pone.0006329. PLoS One. 2009. PMID: 19629181 Free PMC article. - Bifunctional HDAC Therapeutics: One Drug to Rule Them All?
Smalley JP, Cowley SM, Hodgkinson JT. Smalley JP, et al. Molecules. 2020 Sep 24;25(19):4394. doi: 10.3390/molecules25194394. Molecules. 2020. PMID: 32987782 Free PMC article. Review. - Insights into the activation mechanism of class I HDAC complexes by inositol phosphates.
Watson PJ, Millard CJ, Riley AM, Robertson NS, Wright LC, Godage HY, Cowley SM, Jamieson AG, Potter BV, Schwabe JW. Watson PJ, et al. Nat Commun. 2016 Apr 25;7:11262. doi: 10.1038/ncomms11262. Nat Commun. 2016. PMID: 27109927 Free PMC article.
References
- Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. - PubMed
- Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. - PubMed
- Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous