Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element - PubMed (original) (raw)
Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element
Koh Fujinaga et al. Mol Cell Biol. 2004 Jan.
Abstract
The elongation of transcription is a highly regulated process that requires negative and positive effectors. By binding the double-stranded stem in the transactivation response (TAR) element, RD protein from the negative transcription elongation factor (NELF) inhibits basal transcription from the long terminal repeat of the human immunodeficiency virus type 1 (HIVLTR). Tat and its cellular cofactor, the positive transcription elongation factor b (P-TEFb), overcome this negative effect. Cdk9 in P-TEFb also phosphorylates RD at sites next to its RNA recognition motif. A mutant RD protein that mimics its phosphorylated form no longer binds TAR nor represses HIV transcription. In sharp contrast, a mutant RD protein that cannot be phosphorylated by P-TEFb functions as a dominant-negative effector and inhibits Tat transactivation. These results better define the transition from abortive to productive transcription and thus replication of HIV.
Figures
FIG. 1.
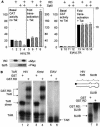
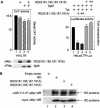
RD and Spt5 inhibit basal transcription and increase Tat transactivation from the HIVLTR but not EIAVLTR. (A) Effects of RD and Spt5 on the HIVLTR in cells. pEF.RD (0.5 μg) and/or pCMV.SPT5 was cotransfected with pHIVCAT (0.1 μg) (lanes 1 to 8) or pEIAVCAT (0.1 μg) (lanes 9 to 12) in the absence or presence of pTat (0.1 μg) or pEIAV-Tat (0.1 μg) into HeLa cells. Two days later, CAT activities were measured. For basal CAT activities, results are presented as raw numbers. For the effects of Tat, results are presented as fold transactivation relative to the CAT activity obtained with the empty plasmid vector (lanes 1 and 9). The expression of RD and Spt5 was visualized by Western blotting using anti-Myc (αMyc) and anti-FLAG (αFlag) antibodies, respectively. The values are means ± standard errors of the mean (error bars) from three independent experiments performed in duplicate. (B) RD binds HIVTAR but not EIAVTAR. 32P-labeled TAR from HIV (lanes 1 and 2), mutant TAR lacking the central loop (Δloop) (lanes 3 and 4), TAR from EIAV (lanes 5 and 6) and SLIIB grafted onto HIVTAR (schematic and lanes 7 and 8) were incubated with (+) purified GST or the GST.RD chimera. The reaction mixture was then separated on a 5% nondenaturing polyacrylamide gel at 4°C. RNA-protein-RNA complexes were visualized by autoradiography. The arrows point to informative RNA-protein complexes, whose composition is given. The position of free probes is indicated by the vertical line at the bottom of the leftmost gel.
FIG. 2.
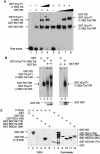
Unphosphorylated RD and the complex between Tat and CycT1 (P-TEFb) bind TAR. (A) RD and the complex between CycT1 and Tat bind TAR simultaneously. In lanes 1 to 4, increasing amounts (0.1 and 0.2 μg for lanes 3 and 4, respectively) of the purified GST.hCycT.Tat chimera (hCycT, human CycT) (7) was added to the reaction mixture containing the 32P-labeled HIVTAR and the GST.RD fusion protein. Purified GST protein (0.2 μg) was used as the negative control (lane 2). In lanes 5 to 8, increasing amounts (0.2, 0.5, and 1.0 μg for lanes 2, 3, and 4, respectively) of the purified GST.RD chimera was added to the reaction mixture containing the 32P-labeled HIVTAR and the GST.hCycT.Tat fusion protein (0.1 μg). Purified GST protein (0.2 μg) was used as the negative control (lane 5). The composition of RNA-protein complexes is given next to the arrows pointing to singly shifted and supershifted bands. This EMSA was visualized by the phosphorimager. (B) Phosphorylated RD does not bind TAR in vitro. GST and the GST.RD chimera were phosphorylated by P-TEFb using cold ATP in vitro. As described above, 0.5 μg of unphosphorylated (lanes 1 to 4) and phosphorylated GST.RD (GST.RDP) chimeras (lanes 5 and 6) were used in the absence or presence (+) of the GST.hCycT.Tat fusion protein (0.1 μg) for the EMSA. Phosphorylation was monitored by a parallel experiment using [γ-32P]ATP (lower right panel). After SDS-PAGE, gels were stained with Coomassie brilliant blue to reveal the levels of input proteins (lower left panel). (C) Sequences N terminal to the RRM in RD are phosphorylated by P-TEFb. (Left) RD is phosphorylated by P-TEFb. GST, GST.RD chimera, and its mutant truncated counterparts [GST.RD(1-106), GST.RD(106-174), GST.RD(174-255), and GST.RD(251-384)] were incubated with P-TEFb and [γ-32P]ATP. Proteins were separated by SDS-PAGE, followed by autoradiography. (Right) The input levels of proteins were verified as described for panel B.
FIG. 3.
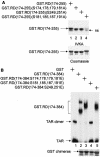
Mapping of the phosphorylation site(s) in RD that is required for dissociating RD and TAR. (A) Mutant RD proteins that were examined in this experiment. IVKA was performed as described in the legend to Fig. 2A using the mutant GST.RD chimeras and P-TEFb. ns, nonspecific band. (B) The abilities of the mutant GST.RD fusion proteins to bind TAR were examined by EMSAs. See the legend to Fig. 2B.
FIG. 4.
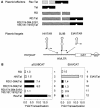
RD binds TAR in vivo. (A) Schematic presentation of plasmid effectors and targets. pA, polyadenylation site. (B) RD can recruit Tat in the RD.Tat chimera to TAR. pEF.RD, pEF.Tat, pEF.RD.Tat, or mutant pEF.RD.Tat plasmid (0.5 μg) was cotransfected with pSLIIBCAT (left) or pEIAVCAT (right) into HeLa cells. CAT activities were measured as described in the legend to Fig. 1A. Results are presented as fold transactivation over the value obtained with the empty plasmid vector.
FIG. 5.
Effects of mutant RD proteins on viral transcription and their incorporation into NELF. (A) (Left) The mutant glutamate-substituted RD protein no longer inhibits HIV transcription. pHIVCAT, mutant RD(S181,185,187,191E) and Spt5 were expressed in HeLa cells, and CAT assays were performed as described in the legend to Fig. 1A. (Right) The mutant alanine-substituted RD protein inhibits Tat transactivation. Tat (0.1 μg), Spt5 (0.1 μg), mutant RD(S181,185,187,191E) (0.1 and 0.3 μg) or mutant RD(S181,185,187,191A) (0.1 and 0.3 μg) proteins were expressed in HeLa cells that stably contained the HIVLTR linked to the luciferase reporter gene. Luciferase activities were measured 48 h later. (B) Wild-type and mutant RD proteins are incorporated equivalently into NELF. 293T cells expressed Myc epitope-tagged RD proteins. Immunoprecipitations (IP) with the anti-NELF-A monoclonal antibody (αNELF-A)were followed by Western blotting (WB) with the anti-Myc antiserum (αMyc). The bottom gel contains 5% of input proteins. In lane 1, cells were transfected with the empty plasmid vector. Arrows point to wild-type and mutant RD proteins in lanes 2, 3, and 4.
FIG. 6.
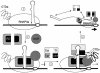
A model for negative and positive transcriptional regulation on the HIVLTR. In step 1, the unphosphorylated RNAPIIa clears the viral promoter. In step 2, the transcription complex is arrested near TAR with the help of DSIF and NELF. RD in NELF binds the lower stem in TAR. After the synthesis of Tat, P-TEFb is recruited to the 5′ bulge and central loop in TAR. Cdk9 phosphorylates the CTD of RNAPIIa (RNAPIIo), Spt5 in DSIF, and RD in NELF (step 3). The phosphorylated RD no longer binds TAR. In step 4, negative factors are converted into positive elongation factors, and RNAPIIo leaves the HIVLTR so that the viral genome is copied efficiently and cotranscriptional processing can take place. Altered N-TEF most likely stays associated with RNAPIIo.
Similar articles
- Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription.
Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Zhou M, et al. J Virol. 2004 Dec;78(24):13522-33. doi: 10.1128/JVI.78.24.13522-13533.2004. J Virol. 2004. PMID: 15564463 Free PMC article. - A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat.
Brès V, Gomes N, Pickle L, Jones KA. Brès V, et al. Genes Dev. 2005 May 15;19(10):1211-26. doi: 10.1101/gad.1291705. Genes Dev. 2005. PMID: 15905409 Free PMC article. - The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation.
Wu SY, Chiang CM. Wu SY, et al. J Biol Chem. 2007 May 4;282(18):13141-5. doi: 10.1074/jbc.R700001200. Epub 2007 Feb 28. J Biol Chem. 2007. PMID: 17329240 Review. - Inhibitors of HIV-1 Tat-mediated transactivation.
Richter SN, Palù G. Richter SN, et al. Curr Med Chem. 2006;13(11):1305-15. doi: 10.2174/092986706776872989. Curr Med Chem. 2006. PMID: 16712471 Review.
Cited by
- Cocaine promotes both initiation and elongation phase of HIV-1 transcription by activating NF-κB and MSK1 and inducing selective epigenetic modifications at HIV-1 LTR.
Sahu G, Farley K, El-Hage N, Aiamkitsumrit B, Fassnacht R, Kashanchi F, Ochem A, Simon GL, Karn J, Hauser KF, Tyagi M. Sahu G, et al. Virology. 2015 Sep;483:185-202. doi: 10.1016/j.virol.2015.03.036. Epub 2015 May 15. Virology. 2015. PMID: 25980739 Free PMC article. - The Role of RNA Polymerase II Elongation Control in HIV-1 Gene Expression, Replication, and Latency.
Nilson KA, Price DH. Nilson KA, et al. Genet Res Int. 2011;2011:726901. doi: 10.4061/2011/726901. Epub 2011 Oct 13. Genet Res Int. 2011. PMID: 22567366 Free PMC article. - Ubiquitylation of Cdk9 by Skp2 facilitates optimal Tat transactivation.
Barboric M, Zhang F, Besenicar M, Plemenitas A, Peterlin BM. Barboric M, et al. J Virol. 2005 Sep;79(17):11135-41. doi: 10.1128/JVI.79.17.11135-11141.2005. J Virol. 2005. PMID: 16103164 Free PMC article. - Runx1 binds positive transcription elongation factor b and represses transcriptional elongation by RNA polymerase II: possible mechanism of CD4 silencing.
Jiang H, Zhang F, Kurosu T, Peterlin BM. Jiang H, et al. Mol Cell Biol. 2005 Dec;25(24):10675-83. doi: 10.1128/MCB.25.24.10675-10683.2005. Mol Cell Biol. 2005. PMID: 16314494 Free PMC article. - Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure.
Mbonye U, Karn J. Mbonye U, et al. Virology. 2014 Apr;454-455:328-39. doi: 10.1016/j.virol.2014.02.008. Epub 2014 Feb 22. Virology. 2014. PMID: 24565118 Free PMC article. Review.
References
- Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. - PMC - PubMed
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
- Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. - PubMed
- Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous