Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1 - PubMed (original) (raw)
Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1
Michelle A Christensen et al. Mol Cell Biol. 2004 Jan.
Abstract
Proteolytic processing of the beta-amyloid precursor protein (APP) at the beta site is essential to generate Abeta. BACE1, the major beta-secretase involved in cleaving APP, has been identified as a type 1 membrane-associated aspartyl protease. We have cloned a 2.1-kb fragment upstream of the human BACE1 gene and identified key regions necessary for promoter activity. BACE1 gene expression is controlled by a TATA-less promoter. The region of bp -619 to +46 is the minimal promoter to control the transcription of the BACE1 gene. Several putative cis-acting elements, such as a GC box, HSF-1, a PU box, AP1, AP2, and lymphokine response element, are found in the 5' flanking region of the BACE1 gene. Transcriptional activation and gel shift assays demonstrated that the BACE1 promoter contains a functional Sp1 response element, and overexpression of the transcription factor Sp1 potentiates BACE gene expression and APP processing to generate Abeta. Furthermore, Sp1 knockout reduced BACE1 expression. These results suggest that BACE1 gene expression is tightly regulated at the transcriptional level and that the transcription factor Sp1 plays an important role in regulation of BACE1 to process APP generating Abeta in Alzheimer's disease.
Figures
FIG. 1.
Sequence features of the human BACE1 gene promoter. (A) Nucleotide sequence of the human BACE1 gene promoter. The 2,668-bp fragment of the 5′ flanking region and the first exon of the human BACE1 gene were isolated from the human genomic library and sequenced by the primer walking strategy. The adenine +1 represents the transcription start site. The positions of some of the unique and common restriction enzymes are in italics. The putative transcription factor binding sites are underlined and in boldface. The codon of the first exon is also indicated. The GenBank accession number is AY162468. (B) Primer extension assay. A primer extension experiment was used to map the BACE1 gene transcription start site. Neuronal RNA was extracted by TRI reagent, and yeast tRNA was used as a control. A 32P-labeled primer complementary to +46 to +24 was used for both primer extension and the sequencing reaction. The DNA fragment of bp −619 to +46 was used as the sequencing template. The samples were analyzed by 6% denaturing PAGE. *, major transcription start site.
FIG. 2.
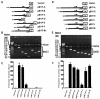
Functional deletion analysis of the human BACE1 gene promoter. (A and D) Schematic diagrams of the BACE1 promoter deletion constructs consisting of a 5′ flanking region with serial deletions cloned into the promoter-less vector plasmid pGL3-basic in front of a reporter gene, the luciferase gene (Luc). Arrow, direction of transcription. The numbers represent the end points of each construct. (B and E) The deletion plasmids shown in panels C and F, respectively, were confirmed by sequencing and restriction enzyme digestion checking, and the digested samples were analyzed on a 1.1% agarose gel. Vector size is 4.7 kb, and the BACE1 gene 5′ flanking fragment insert sizes range from 0.4 to 2.2 kb. (C and F). The constructed plasmids were cotransfected into HEK293T cells with pCH110. Luciferase activity was measured at 48 h by a luminometer. β-Gal activity was used to normalize transfection efficiency. The values represent means ± standard errors of the means (n = 3 to 6). *, P < 0.001 by ANOVA with the post hoc Newmann-Keuls test.
FIG. 3.
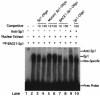
Gel mobility shift assay for the BACE1 gene promoter. Gel shift and gel supershift assays were performed as described in Materials and Methods with the 32P-labeled double-stranded oligonucleotide probe BACE1-Sp1. Lane 1, labeled probe without nuclear extract. Incubation of 32P-labeled BACE1-Sp1 with HeLa nuclear extracts retarded the migration rate of the labeled probe, which formed a new shifted DNA-protein complex band (lane 2). Competition assays were performed by further adding different concentrations of molar excess of unlabeled competition oligonucleotides, consensus Sp1 (lanes 3 and 4), mutant consensus Sp1 (lanes 5 and 6), and homologous BACE1-Sp1 (lanes 7 and 8). The anti-Sp1 antibody was used for the gel supershift assay. The anti-Sp1 antibody supershifted the nucleoprotein-BACE1-Sp1 complex (lane 9), and incubation of the unlabeled consensus Sp1 oligonucleotide competitor abolished the shifted and supershifted bands (lane 10).
FIG. 4.
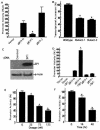
Sp1 binding site is required for the BACE1 promoter function, and the transcription factor Sp1 facilitates the BACE1 gene transcriptional activation. (A) PC12 cells were transfected with plasmids pB1P-H, pB1P-I, and pB1P-J and the empty vector control. The fragment of bp −932 to +292 or bp −896 to +292 from the BACE1 promoter was cloned into pGL3-basic to generate plasmids containing or not containing the Sp1 binding site, pB1P-H and pB1P-I, respectively. Luciferase activity was measured at 72 h by a luminometer. β-Gal activity was used to normalize transfection efficiency. The values represent means ± standard errors of the means (SEM) (n = 3 to 6). *, P < 0.001 relative to pB1P-H and pB1P-J by ANOVA with post hoc Newmann-Keuls test. (B) Plasmids pB1P-H (wild type), pB1P-H-mut1Sp1 (Mutant-1), and pB1P-H-mut1Sp1 (Mutant-2) were transfected into HEK293T cells, and the promoter activity was measured. Two Sp1 binding site mutations significantly reduced the BACE1 promoter activity (n = 3; *, P < 0.001). (C) Western blot detection of Sp1 in control cells transfected with empty vector and cells transfected with Sp1 expression plasmid pCGN-Sp1. Ten micrograms of cell lysates from cells transfected for 48 h was analyzed by 4 to 20% Tris-glycine SDS-PAGE. The Sp1 protein was robustly expressed in pCGN-Sp1-transfected cells, and β-actin was used as an internal protein control. (D) Transcriptional activation of the BACE1 promoter is potentiated by Sp1. The empty vector, the Sp1-binding site containing BACE1 promoter plasmid pB1P-H, and the BACE1 promoter plasmid lacking the Sp1 binding site, pB1P-I, were cotransfected with Sp1 expression plasmid pCGN-Sp1 into cells. Overexpression of Sp1 significantly increased the pB1P-H BACE1 promoter activity by over fourfold and had no significant effect on pB1P-I and the control plasmid (n = 3; *, P < 0.001). (E and F). Inhibition of the BACE1 promoter activity by mithramycin A. The BACE1 promoter construct pB1P-H was transfected in HEK293T cells. The transfected cells were treated with vehicle solution control or mithramycin A for 48 h at 25, 75, or 125 nM (E) for the dosage-dependent assay or with mithramycin A at 125 nM for 16 or 48 h for the time course assay (F). Cells were harvested at the same transfection end point, and luciferase activity was measured and expressed as means ± SEM relative to control promoter activity. *, P < 0.01 relative to control by ANOVA with post hoc Newmann-Keuls test.
FIG. 5.
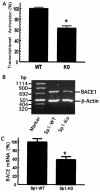
BACE1 gene transcription is markedly reduced in Sp1-KO cells. (A) Sp1-induced transcriptional activation of the BACE1 promoter is markedly reduced in Sp1-KO cells. Sp1-WT and Sp1-KO cells were cotransfected with the BACE1 promoter plasmid pB1P-H and the β-Gal expression plasmid. Values represent the percentages of normalized luciferase activity and represent the means ± standard errors of the means (SEM) (*, P < 0.01 relative to Sp1-WT cells by ANOVA with the post hoc Student Newmann-Keuls test. (B) Endogenous BACE1 mRNA level is reduced in Sp1-KO cells. RNA was isolated from Sp1-WT and Sp1-KO cells. Quantitative RT-PCR was performed to measure the endogenous level of the BACE1 mRNA to assay BACE1 gene transcription in vivo. Specific BACE1 and β-actin coding sequence primers were used to amplify the BACE1 and β-actin cDNA, as described in Materials and Methods. Different cycles and amounts of PCR products were analyzed, and the DNA gel represents 25 cycles of RT-PCR products on 1.2% agarose gel. (C) The ratio of BACE1 to β-actin gene transcription in Sp1-WT (WT) and Sp1-KO (KO) cells was quantitated by Kodak Image Analysis. The endogenous BACE1 mRNA level was significantly decreased in Sp1-KO cells relative to that in Sp1-WT cells. Shown are the means ± SEM (*, P < 0.01 relative to Sp1-WT cells by t test).
FIG. 6.
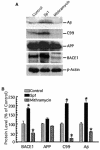
Sp1 potentiates Aβ generation by upregulating BACE1 activity. (A) HEK293T cells stably expressing Swedish mutant APP were transfected with the pCGN-Sp1 plasmid (Sp1) or not transfected (control) and treated with mithramycin A at 125 nM (mithramycin) for 48 h. Western blot analysis was performed to detect Aβ, APP C99, full-length APP, BACE1, and β-actin. A monoclonal anti-β-actin antibody (AC-15) was used to detect β-actin, and the BACE1 protein level was detected by rabbit polyclonal BACE1 antibody LK-16. To detect APP C99, the major BACE1 cleavage product, cell lysates were analyzed by 10 to 20% Tris-Tricine gel with the 6E10 antibody. For Aβ detection, conditioned media were first immunoprecipitated with Aβ antibody 4G8 and an immunoblot assay was then performed to analyze the precipitates on 10 to 20% Tris-Tricine gel with monoclonal antibody 6E10. Note that overexpression of Sp1 increases BACE1 protein and subsequently affects APP processing at the β site, while inhibition of Sp1 binding by mithramycin A has the opposite effect. No significant changes in β-actin and full-length APP levels were detected. (B) Quantitative analysis of the generation of Aβ, APP C99, full-length APP, and BACE1. Values are means ± standard errors of the means (n = 3). The protein levels are expressed as percentages of the levels in control cells. *, P < 0.01 relative to controls by ANOVA with post hoc Newmann-Keuls test.
Similar articles
- Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes.
Sun X, Wang Y, Qing H, Christensen MA, Liu Y, Zhou W, Tong Y, Xiao C, Huang Y, Zhang S, Liu X, Song W. Sun X, et al. FASEB J. 2005 May;19(7):739-49. doi: 10.1096/fj.04-3426com. FASEB J. 2005. PMID: 15857888 - BACE1 gene expression and protein degradation.
Zhou W, Qing H, Tong Y, Song W. Zhou W, et al. Ann N Y Acad Sci. 2004 Dec;1035:49-67. doi: 10.1196/annals.1332.004. Ann N Y Acad Sci. 2004. PMID: 15681800 - Functional characterization of the 5' flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression.
Ge YW, Maloney B, Sambamurti K, Lahiri DK. Ge YW, et al. FASEB J. 2004 Jun;18(9):1037-9. doi: 10.1096/fj.03-1379fje. Epub 2004 Apr 1. FASEB J. 2004. PMID: 15059977 - Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease.
Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Rossner S, et al. Prog Neurobiol. 2006 Jun;79(2):95-111. doi: 10.1016/j.pneurobio.2006.06.001. Epub 2006 Aug 14. Prog Neurobiol. 2006. PMID: 16904810 Review. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
Cited by
- Early Growth Response 1 (Egr-1) Is a Transcriptional Activator of β-Secretase 1 (BACE-1) in the Brain.
Qin X, Wang Y, Paudel HK. Qin X, et al. J Biol Chem. 2016 Oct 14;291(42):22276-22287. doi: 10.1074/jbc.M116.738849. Epub 2016 Aug 30. J Biol Chem. 2016. PMID: 27576688 Free PMC article. - Amyloid Beta-Peptide Increases BACE1 Translation through the Phosphorylation of the Eukaryotic Initiation Factor-2_α_.
Picón-Pagès P, Gutiérrez DA, Barranco-Almohalla A, Crepin G, Tajes M, Ill-Raga G, Guix FX, Menéndez S, Arumí-Uría M, Vicente R, Álvarez AR, Muñoz FJ. Picón-Pagès P, et al. Oxid Med Cell Longev. 2020 Sep 19;2020:2739459. doi: 10.1155/2020/2739459. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33014268 Free PMC article. - BACE1 gene promoter single-nucleotide polymorphisms in Alzheimer's disease.
Zhou W, Cai F, Li Y, Yang GS, O'Connor KD, Holt RA, Song W. Zhou W, et al. J Mol Neurosci. 2010 Sep;42(1):127-33. doi: 10.1007/s12031-010-9381-6. Epub 2010 May 9. J Mol Neurosci. 2010. PMID: 20455082 - Deficiency of Neuronal p38α MAPK Attenuates Amyloid Pathology in Alzheimer Disease Mouse and Cell Models through Facilitating Lysosomal Degradation of BACE1.
Schnöder L, Hao W, Qin Y, Liu S, Tomic I, Liu X, Fassbender K, Liu Y. Schnöder L, et al. J Biol Chem. 2016 Jan 29;291(5):2067-79. doi: 10.1074/jbc.M115.695916. Epub 2015 Dec 9. J Biol Chem. 2016. PMID: 26663083 Free PMC article. - Tripchlorolide Attenuates β-amyloid Generation via Suppressing PPARγ-Regulated BACE1 Activity in N2a/APP695 Cells.
Lin N, Chen LM, Pan XD, Zhu YG, Zhang J, Shi YQ, Chen XC. Lin N, et al. Mol Neurobiol. 2016 Nov;53(9):6397-6406. doi: 10.1007/s12035-015-9542-2. Epub 2015 Nov 19. Mol Neurobiol. 2016. PMID: 26582466
References
- Basler, K., B. Oesch, M. Scott, D. Westaway, M. Walchli, D. F. Groth, M. P. McKinley, S. B. Prusiner, and C. Weissmann. 1986. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell 46:417-428. - PubMed
- Benjannet, S., A. Elagoz, L. Wickham, M. Mamarbachi, J. S. Munzer, A. Basak, C. Lazure, J. A. Cromlish, S. Sisodia, F. Checler, M. Chretien, and N. G. Seidah. 2001. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 276:10879-10887. - PubMed
- Bennett, B. D., P. Denis, M. Haniu, D. B. Teplow, S. Kahn, J. C. Louis, M. Citron, and R. Vassar. 2000. A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer's beta-secretase. J. Biol. Chem. 275:37712-37717. - PubMed
- Beyreuther, K., T. Dyrks, C. Hilbich, U. Monning, G. Konig, G. Multhaup, P. Pollwein, and C. L. Masters. 1992. Amyloid precursor protein (APP) and beta A4 amyloid in Alzheimer's disease and Down syndrome. Prog. Clin. Biol. Res. 379:159-182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous