In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding - PubMed (original) (raw)
In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding
Brigitte D Lavoie et al. Genes Dev. 2004.
Erratum in
- Genes Dev. 2004 Feb 1;18(3):355
Abstract
Chromosome condensation plays an essential role in the maintenance of genetic integrity. Using genetic, cell biological, and biochemical approaches, we distinguish two cell-cycle-regulated pathways for chromosome condensation in budding yeast. From G(2) to metaphase, we show that the condensation of the approximately 1-Mb rDNA array is a multistep process, and describe condensin-dependent clustering, alignment, and resolution steps in chromosome folding. We functionally define a further postmetaphase chromosome assembly maturation step that is required for the maintenance of chromosome structural integrity during segregation. This late step in condensation requires the conserved mitotic kinase Ipl1/aurora in addition to condensin, but is independent of cohesin. Consistent with this, the late condensation pathway is initiated during the metaphase-to-anaphase transition, supports de novo condensation in cohesin mutants, and correlates with the Ipl1/aurora-dependent phosphorylation of condensin. These data provide insight into the molecular mechanisms of higher-order chromosome folding and suggest that two distinct condensation pathways, one involving cohesins and the other Ipl1/aurora, are required to modulate chromosome structure during mitosis.
Figures
Figure 1.
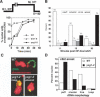
Cell-cycle-dependent intermediates in rDNA condensation. (A) A cdc15-1 (YBL26c-9a) strain was used to synchronize the cells in M phase. Following release to the permissive temperature to monitor an unperturbed G1, nocodazole was added to the culture 1 h postrelease to prevent cycling beyond the first cell cycle. Time points were taken as shown, and cells were processed for flow cytometric analysis and FISH, using an rDNA probe. (B) Micrographs of FISH of the yeast rDNA (FITC, green) and chromosomes (PI, red). (Bottom panels) The isolated rDNA signal. At each time point, a representative micrograph of the most prominent or newly emerging species is shown. Letters classify the morphologies detected, the quantitation of which is shown in C. At least 100 nuclei were scored per time point.
Figure 2.
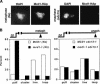
Condensation intermediates following mitotic arrest. (A) Kinetics of chromosome condensation in M phase. An experimental schematic is shown. Strains CH2523 (WT), CH2524 (brn1-9), and 2153-1 (ycg1-2) were synchronized in G1 with α-factor, shifted to 37°C (0.5 h), and rearrested in mitosis following release into nocodazole-containing media. The cultures were returned to the permissive temperature (23°C), and samples were taken as indicated. rDNA FISH was performed, and the percent nuclei displaying rDNA loops were scored. (B) Quantitation of rDNA species during mitotic condensation. Strain 2153-1 (ycg1-2) was treated as in A, and condensation intermediates were scored by FISH at 0, 15, and 30 min after release to the permissive temperature. The percent nuclei with puff, cluster, line, or loop rDNA morphologies are plotted at each time point. At least 100 nuclei were scored per sample. (C) Accumulation of clusters in a ycg1-2 mutant. Strains YPH499 (WT), YBL02–18b (integrated ycg1-2), and 2125–3a/pBL238-ts2 (ycg1::KAN/CEN6 ARS1 LEU2 ycg1-2) were synchronized in G1, shifted to the restrictive temperature, released, and rearrested in mitosis with nocodazole, followed by rDNA FISH to determine rDNA morphology. Chromosomes are depicted in red with the rDNA signal in green/yellow. (D) Postanaphase rDNA clusters and lines are condensin-dependent. Strains 982–101 (YCG1 GAL1-CLB2_Δ_db) and 2172–101 (ycg1-2 GAL1-CLB2_Δ_db) were arrested in late M by overexpression of undegradable Clb2p (see Materials and Methods). The cells were then shifted to the restrictive temperature for 30 min, fixed, and processed for rDNA FISH. Quantitation of rDNA morphologies is shown. More than 100 nuclei were counted per sample.
Figure 3.
Chromosome condensation in the absence of Mcd1p. (A) Chromatin spreads of Mcd1-HAp in metaphase-versus-anaphase cells. YBL13–2a (MCD1-HA6 cdc15-1) cells were synchronized in G1, released, and rearrested at 37°C either with nocodazole or by allowing the cells to proceed to the cdc15-1 block. Mcd1-HAp was monitored by indirect immunofluorescence using an anti-HA monoclonal antibody (16B12; see Materials and Methods). (B) rDNA condensation in metaphase-versus-anaphase-arrested cells. Cultures of YBL26–5–3a (MCD1 cdc15-1) and YBL111 (mcd1-1 cdc15-1) were synchronized in G1, shifted to the restrictive temperature, and released into prewarmed media plus or minus nocodazole as indicated. Following rearrest in metaphase or anaphase (as indicated), rDNA FISH was performed. Quantitation of rDNA puffs, clusters, lines, and loops is shown. More than 100 nuclei were scored per sample.
Figure 4.
Mitotic phosphorylation of the condensin subunit Ycg1p. (A) Western blot analysis of Ycg1–HA3p (2131a) from asynchronously growing (Asy) and synchronized cells (G1, S, and M phase). Whole-cell extracts were separated by SDS-PAGE (6.5% resolving gel), transferred to nitrocellulose, and probed with a mouse monoclonal anti-HA antibody (16B12), which recognizes a doublet of Ycg1p (arrows) and an unrelated 125-kD polypeptide just below. The slower migrating form is sensitive to alkaline phosphatase treatment (right panel). Ycg1–HAp was immunoprecipitated from nocodazole-arrested cells using anti-HA antibodies and protein A Sepharose, and treated plus and minus alkaline phosphatase. Arrows indicate both forms of Ycg1–HAp. (B) Western blot of Ycg1–HAp phosphorylation in IPL1+ (YBL55–9b) and ipl1-321 (YBL55–10b) strains. Cells were synchronized in G1, shifted to 37°C, and released into warm medium containing nocodazole to rearrest the cells in mitosis. G1-versus-M-phase extracts are shown. Arrows mark Ycg1p phospho-forms. (C) Correlation of Ycg1p phosphorylation with rDNA condensation. Following cell synchronization in a cdc15-1 background, cells were released into the cell cycle at 23°C in the presence of nocodazole, and aliquots were taken at each time point for rDNA FISH and FACS analysis (see Fig. 1) as well as for protein analysis by Western blot. Under- and overphosphorylated forms of Ycg1p are indicated (arrows). The amount of Ycg1p shifted to the slower migrating band is plotted over time (lower panel). The temporal succession of rDNA species is redrawn from Fig. 1 for comparison.
Figure 5.
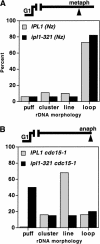
Ipl1 kinase requirement for chromosome structure postanaphase onset. FISH analysis of rDNA condensation in IPL1 (YBL26–5–3a; IPL1 cdc15-1) versus ipl1-321 (YBL76–9b; ipl1-321 cdc15-1) strains in metaphase (A) versus anaphase cells (B). Following synchronization in G1, cells were released at 37°C and rearrested in metaphase using nocodazole (A) or allowed to proceed to the anaphase block (cdc15-1; B). After 2.5 h, cells were fixed and processed for rDNA FISH. rDNA morphologies were scored as indicated. More than 100 nuclei were counted per sample.
Figure 6.
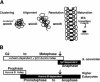
Condensation is a multistep process and can proceed through two distinct pathways. (A) Model of chromosome condensation. As cells enter into the G2/early M phase of the cell cycle, condensation of the chromosomes proceeds through three distinct stages: clustering, alignment, and resolution. Both condensins (gray balls) and cohesins (black bars) are required for the establishment and maintenance of condensation until metaphase. Resolution of the more compacted cluster to lines and finally loops could result from protein–protein interactions between condensin molecules, in a manner reminiscent of higher-order nucleoprotein assembly pathways (see Discussion). At the metaphase-to-anaphase transition, however, a different condensation pathway operates. The Ipl1 kinase promotes a further maturation of chromosomes necessary for the maintenance of structural integrity in the absence of cohesins. This maturation event may in part be accomplished through the phosphorylation of condensin, namely, through its Ycg1p subunit. Condensin would thus have two distinct active forms specific to each condensation pathway: a foldase that promotes cohesin-dependent condensation and an Ipl1-dependent maturase required postanaphase onset. (B) Conservation of condensation pathways in budding yeast versus higher eukaryotes. In S. cerevisiae, the Ipl1/aurora-independent (cohesin-dependent) pathway is activated in G2/M and persists to metaphase (white bar). Prior to the metaphase-to-anaphase transition (indicated by an arrow), the Ipl1/aurora kinase pathway (black bar) is activated, such that chromosome structural integrity is maintained during anaphase. In higher eukaryotes, condensin function initiates in prophase in an aurora-independent manner and during which time cohesins are still present on chromosomes. In late prophase, aurora stimulates the loading of condensin on chromatin, coincident with the removal of bulk cohesins, and suggesting that the Ipl1/aurora pathway is activated significantly before the metaphase-to-anaphase transition.
Similar articles
- In vivo analysis of chromosome condensation in Saccharomyces cerevisiae.
Vas AC, Andrews CA, Kirkland Matesky K, Clarke DJ. Vas AC, et al. Mol Biol Cell. 2007 Feb;18(2):557-68. doi: 10.1091/mbc.e06-05-0454. Epub 2006 Dec 6. Mol Biol Cell. 2007. PMID: 17151360 Free PMC article. - Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase.
Sullivan M, Higuchi T, Katis VL, Uhlmann F. Sullivan M, et al. Cell. 2004 May 14;117(4):471-82. doi: 10.1016/s0092-8674(04)00415-5. Cell. 2004. PMID: 15137940 - Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity.
St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauvé V, Ratsima H, D'Amours D. St-Pierre J, et al. Mol Cell. 2009 May 14;34(4):416-26. doi: 10.1016/j.molcel.2009.04.013. Mol Cell. 2009. PMID: 19481522 - Three-step model for condensin activation during mitotic chromosome condensation.
Bazile F, St-Pierre J, D'Amours D. Bazile F, et al. Cell Cycle. 2010 Aug 15;9(16):3243-55. doi: 10.4161/cc.9.16.12620. Epub 2010 Aug 7. Cell Cycle. 2010. PMID: 20703077 Review. - Protein kinases in mitotic phosphorylation of budding yeast CENP-A.
Mishra PK, Basrai MA. Mishra PK, et al. Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review.
Cited by
- Nutrient starvation promotes condensin loading to maintain rDNA stability.
Tsang CK, Li H, Zheng XS. Tsang CK, et al. EMBO J. 2007 Jan 24;26(2):448-58. doi: 10.1038/sj.emboj.7601488. Epub 2007 Jan 4. EMBO J. 2007. PMID: 17203076 Free PMC article. - Cohesin: an emerging master regulator at the heart of cardiac development.
Mfarej MG, Hyland CA, Sanchez AC, Falk MM, Iovine MK, Skibbens RV. Mfarej MG, et al. Mol Biol Cell. 2023 May 1;34(5):rs2. doi: 10.1091/mbc.E22-12-0557. Epub 2023 Mar 22. Mol Biol Cell. 2023. PMID: 36947206 Free PMC article. - Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis.
Resnick TD, Dej KJ, Xiang Y, Hawley RS, Ahn C, Orr-Weaver TL. Resnick TD, et al. Genetics. 2009 Mar;181(3):875-87. doi: 10.1534/genetics.108.097741. Epub 2008 Dec 22. Genetics. 2009. PMID: 19104074 Free PMC article. - Insights into dynamic mitotic chromatin organization through the NIMA kinase suppressor SonC, a chromatin-associated protein involved in the DNA damage response.
Larson JR, Facemyer EM, Shen KF, Ukil L, Osmani SA. Larson JR, et al. Genetics. 2014 Jan;196(1):177-95. doi: 10.1534/genetics.113.156745. Epub 2013 Nov 8. Genetics. 2014. PMID: 24214344 Free PMC article. - EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases.
Kapoor P, Lavoie BD, Frappier L. Kapoor P, et al. Mol Cell Biol. 2005 Jun;25(12):4934-45. doi: 10.1128/MCB.25.12.4934-4945.2005. Mol Cell Biol. 2005. PMID: 15923612 Free PMC article.
References
- Amon A., Irniger, S., and Nasmyth, K. 1994. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell 77: 1037–1050. - PubMed
- Bazett-Jones D.P., Kimura, K., and Hirano, T. 2002. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9: 1183–1190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous