Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation - PubMed (original) (raw)
. 2004 Jan 1;18(1):99-115.
doi: 10.1101/gad.276304. Epub 2003 Dec 30.
Affiliations
- PMID: 14701881
- PMCID: PMC314285
- DOI: 10.1101/gad.276304
Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation
Luika A Timmerman et al. Genes Dev. 2004.
Abstract
Epithelial-to-mesenchymal transition (EMT) is fundamental to both embryogenesis and tumor metastasis. The Notch intercellular signaling pathway regulates cell fate determination throughout metazoan evolution, and overexpression of activating alleles is oncogenic in mammals. Here we demonstrate that Notch activity promotes EMT during both cardiac development and oncogenic transformation via transcriptional induction of the Snail repressor, a potent and evolutionarily conserved mediator of EMT in many tissues and tumor types. In the embryonic heart, Notch functions via lateral induction to promote a selective transforming growth factor-beta (TGFbeta)-mediated EMT that leads to cellularization of developing cardiac valvular primordia. Embryos that lack Notch signaling elements exhibit severely attenuated cardiac snail expression, abnormal maintenance of intercellular endocardial adhesion complexes, and abortive endocardial EMT in vivo and in vitro. Accordingly, transient ectopic expression of activated Notch1 (N1IC) in zebrafish embryos leads to hypercellular cardiac valves, whereas Notch inhibition prevents valve development. Overexpression of N1IC in immortalized endothelial cells in vitro induces EMT accompanied by oncogenic transformation, with corresponding induction of snail and repression of VE-cadherin expression. Notch is expressed in embryonic regions where EMT occurs, suggesting an intimate and fundamental role for Notch, which may be reactivated during tumor metastasis.
Figures
Figure 1.
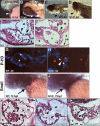
Endocardial expression of Notch and marker genes. Whole-mount and transverse sections of E8.5–9.5 hearts. (A–D) Delta4 mRNA is expressed in the endocardium of wild-type embryos (A, C), and is severely reduced in RBPJk mutants (B, D). (E) Semiquantitative RT-PCR analysis in E8.5–9.0 wild-type and RBPJk mutants. (B) Body; (H) heart. (F–I) Notch1 is strongly expressed in the endocardium of wild-type embryos (F, H) and is strongly reduced in RBPJk mutants (G, I). (J–M) Endocardial HRT1 expression in wild-type (J, L) is strongly reduced in RBPJk mutants (K, M). (N–Q) PECAM1protein is present in the endocardium of wild-type (N, P) and RBPJk (O, Q) embryos. (R–U) GATA5 is expressed in the endocardium and myocardium of wild-type (R, T) and RBPJk (S, U) embryos. (V–Z) E9.5 wild-type hearts. (V, X, Y) Notch1 expression. (_V, Z, Z_′) Double in situ with Delta4 (red) and Notch1 (blue). (X, Y) Notch1 expression in endocardium (arrow in Y) and mesenchyme (arrowhead in Y); (_Z, Z_′) Delta4 and Notch1 coexpression. Coincident signal produces black/purple coloration. Dashed lines indicate plane of section. (en) Endocardium; (ot) outflow tract; (avc) atrio-ventricular canal; (at) atrium; (lv) left ventricle, (rv) right ventricle; (mes) mesenchyme. Note the defective looping of RBPJk mutants. Scale bar, 25 μm in M, O; 100 μm in L, N.
Figure 2.
Notch pathway mutants initiate endocardial EMT. (A–B) Diagrams showing a lateral view of E9.5 wild-type (A) and RBPJk mutant (B) hearts with the cardiac chambers depicted in color. The Roman numbers indicate the levels of the sections shown in panels E_–_P. (E–H) H&E-stained E9.5 heart transverse sections at the atrio-ventricular canal level. (E, F) Wild type. (F) Mesenchymal cells (arrowhead) invading the cushion. (G, H) RBPJk mutants collapsed endocardium (arrow). (I–P) TEM of E9.5 atrio-ventricular canal endocardia. (I) Wild-type rounded endocardial cells with processes directed toward the cardiac jelly (arrows), among flattened nonmigratory endothelial cells (arrowhead). (J, L) RBPJk mutant endocardial cells extending pseudopodia (arrow in L). Lipid vesicles (asterisks) are found in RBPJk mutant endocardial cells (K, L) and in wild-type migratory mesenchymal cells (N). Adherens junctions (arrowheads in O, P) are conspicuous in RBPJk mutants. Scale bar, 25 μm in F, H; 100 μm in E, G.
Figure 3.
Reduced TGF_β_2 and Snail expression with persistent VE-cadherin expression associate to an impaired EMT in Notch pathway mutants. E9.5 hearts whole-mount and transverse sections. (A–C) TGF_β_2 expression in wild-type (A), RBPJk (B), and Notch1 (C) mutants. (D) Semiquantitative RT-PCR in E9.5–10.0 embryos; (B) body; (H) heart. (E–G, K, M) snail transcription in E9.5 versus E9.5–10.0 hearts. (E, F, K) Wild type. (G, M) RBPJk mutants. (H–J, L, N) VE-cadherin expression in E9.5 versus E9.5–10.0 hearts. (H, I, L) Wild type. (J, N) RBPJk mutants. Scale bar, 50 μm.
Figure 4.
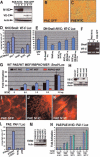
Impaired EMT in Notch1, RBPJk, and DAPT-treated cardiac explants cultures. E9.5 atrio-ventricular canal explants cultured for 72 or 96 h. (A, B) Wild type. (C, D) Notch1 mutant. (E, F) RBPJk mutant. (G, H) Wild-type explant cultured with DMSO. (I, J) Wild-type explant cultured with 10 μM DAPT. (B, D, F, H, J) Phalloidin-TRITC staining. Transformed mesenchymal cells invadingthe collagen gel are indicated by arrowheads, and activated but not transformed cells by arrows. (K) Quantitative analysis of explants. (Left table) The number of transformed mesenchymal cells is significantly reduced in explants from Notch1 and RBPJk mutants. (Right table) Wild-type DAPT-treated explants show a dose-dependent reduction in transformed mesenchymal cells and an increase in activated but not transformed cells. (m) Myocardium. (L) Wild-type explant with transformed cells expressing α-SMA-Cy3 (arrowheads) surrounded by nontransformed endothelial cells stained with Phalloidin-FITC. Scale bar, 15 μm in A, C, E, G, I; 5 μm in B, D, F, H, J; and 2 μm in L.
Figure 5.
Manipulation of Notch signaling activity perturbs valve development in zebrafish. (A, B) In situ hybridization showing mouse N1IC expression in the heart (arrowhead) of 56- and 72-hpf injected zebrafish embryos. (C, D) Embryos at 5 d postfertilization. (C) Wild type. (D) N1IC mRNA-injected embryo exhibiting pericardial edema. Arrowhead points to pericardial sac in C, D. (E, F) H&E sagittal sections; arrowheads indicate the leaflets of the bicuspid atrio-ventricular valves. (E) Wild-type embryo. (F) Representative N1IC-injected embryo with pericardial distention. Note the abnormally large size of the atrio-ventricular valves (arrowheads). The asterisk labels the large pericardial space in the injected embryos. (G, H) Mitotic cells labeled with anti-phospho-histone H3 (red) at 4 d postfertilization in sagittal sections of control-(G) and N1IC-injected (H) embryos. Note the presence of a mitotic cell (arrowhead) in the endocardial cushion of the N1IC-injected embryo (H). DAPI (blue) stains cell nuclei. The position of the atrio-ventricular valve is indicated by asterisks. (I, J) Detection of zebrafish snail1 transcripts by whole-mount in situ hybridization in 3 d postfertilization control-(I) and N1IC-injected (J) embryos. Note the up-regulation of snail1 transcripts in the heart region of N1IC-injected embryos (arrow in J). (K–M) H&E sagittal sections of 5 d postfertilization embryos treated with DMSO (K), or DAPT at final concentrations of 50 (L) or 100 (M) μM. Arrowheads indicate the leaflets of the bicuspid atrio-ventricular valves. Note the valve atrophy in embryos in which Notch activity was down-regulated (K, M). The asterisk labels the pericardial space. In all of the images, embryo view is lateral, dorsal to the top, anterior to the left. (at) Atrium; (pe) pericardium; (v) ventricle. Scale bar, 50 μm.
Figure 6.
Constitutive Notch signaling produces an invasive phenotype via snail induction and VE-cadherin repression in PAE cells. (A) PAE-N1IC clones express little VE-cadherin. (B, C) Cells on plastic (10×). (B) PAE-GFP (wild type). (C) PAE N1IC. (D) Snail and N1IC significantly repress VE-cadherin promoter activity. (E) DN-snail suppresses VE-cadherin repression mediated by N1IC. (F) RT-PCR analysis of snail and VE-cadherin expression in PAE-N1IC clones 1 and 2 and in PAE-GFP clone 3. Snail is induced in N1IC-expressing clones, and VE-cadherin is down-regulated. (G) The snail promoter is activated by N1IC in a dose-dependent manner in wild-type PAE (black bars) and MEFs (blue bars), but not in RBPJk mutant MEFs (red bars). (H) Expression analysis of PAE-snail subclones. (I–K) Cells in collagen I matrix. (I) PAE-GFP (10×). (J, K) PAE N1IC (10×, 60×). (L) Notch1-IC does not induce SMAD activity in PAE cells. (M) PAE cells express different TBRIII levels irrespective of N1IC. (N) Activation of the PAI-Luc reporter correlates with TBRIII expression levels but not with N1IC. (N1IC) myc-epitope tagged N1IC; (VE-C) VE-cadherin; (Actin) β-actin; (Snail) HA epitope-tagged Snail; (p50) NF-kB-p50; (pCDNA3) empty vector; (GAPDH) glyceraldehyde-3-phosphate dehydrogenase.
Similar articles
- Notch and epithelial-mesenchyme transition in development and tumor progression: another turn of the screw.
Grego-Bessa J, Díez J, Timmerman L, de la Pompa JL. Grego-Bessa J, et al. Cell Cycle. 2004 Jun;3(6):718-21. Epub 2004 Jun 28. Cell Cycle. 2004. PMID: 15197341 Review. - Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor.
Saad S, Stanners SR, Yong R, Tang O, Pollock CA. Saad S, et al. Int J Biochem Cell Biol. 2010 Jul;42(7):1115-22. doi: 10.1016/j.biocel.2010.03.016. Epub 2010 Mar 27. Int J Biochem Cell Biol. 2010. PMID: 20348013 - Slug is a direct Notch target required for initiation of cardiac cushion cellularization.
Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Niessen K, et al. J Cell Biol. 2008 Jul 28;182(2):315-25. doi: 10.1083/jcb.200710067. J Cell Biol. 2008. PMID: 18663143 Free PMC article. - The protein tyrosine phosphatase Pez regulates TGFbeta, epithelial-mesenchymal transition, and organ development.
Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y. Wyatt L, et al. J Cell Biol. 2007 Sep 24;178(7):1223-35. doi: 10.1083/jcb.200705035. J Cell Biol. 2007. PMID: 17893246 Free PMC article. - Blockade of Jagged/Notch pathway abrogates transforming growth factor β2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells.
Chen X, Xiao W, Liu X, Zeng M, Luo L, Wu M, Ye S, Liu Y. Chen X, et al. Curr Mol Med. 2014 May;14(4):523-34. doi: 10.2174/1566524014666140331230411. Curr Mol Med. 2014. PMID: 24694299 Review.
Cited by
- Mechanisms of heart valve development and disease.
O'Donnell A, Yutzey KE. O'Donnell A, et al. Development. 2020 Jul 3;147(13):dev183020. doi: 10.1242/dev.183020. Development. 2020. PMID: 32620577 Free PMC article. Review. - Atrioventricular valve development: new perspectives on an old theme.
de Vlaming A, Sauls K, Hajdu Z, Visconti RP, Mehesz AN, Levine RA, Slaugenhaupt SA, Hagège A, Chester AH, Markwald RR, Norris RA. de Vlaming A, et al. Differentiation. 2012 Jul;84(1):103-16. doi: 10.1016/j.diff.2012.04.001. Epub 2012 May 11. Differentiation. 2012. PMID: 22579502 Free PMC article. Review. - Kaposi sarcoma herpesvirus promotes endothelial-to-mesenchymal transition through Notch-dependent signaling.
Gasperini P, Espigol-Frigole G, McCormick PJ, Salvucci O, Maric D, Uldrick TS, Polizzotto MN, Yarchoan R, Tosato G. Gasperini P, et al. Cancer Res. 2012 Mar 1;72(5):1157-69. doi: 10.1158/0008-5472.CAN-11-3067. Epub 2012 Jan 11. Cancer Res. 2012. PMID: 22237624 Free PMC article. - New Concepts in the Development and Malformation of the Arterial Valves.
Henderson DJ, Eley L, Chaudhry B. Henderson DJ, et al. J Cardiovasc Dev Dis. 2020 Sep 24;7(4):38. doi: 10.3390/jcdd7040038. J Cardiovasc Dev Dis. 2020. PMID: 32987700 Free PMC article. Review. - Notch1 phenotype and clinical stage progression in non-small cell lung cancer.
Nguyen D, Rubinstein L, Takebe N, Miele L, Tomaszewski JE, Ivy P, Doroshow JH, Yang SX. Nguyen D, et al. J Hematol Oncol. 2015 Feb 6;8:9. doi: 10.1186/s13045-014-0104-2. J Hematol Oncol. 2015. PMID: 25653136 Free PMC article.
References
- Akhurst R.J., Lehnert, S.A., Faissner, A., and Duffie, E. 1990. TGF beta in murine morphogenetic processes: The early embryo and cardiogenesis. Development 108: 645–656. - PubMed
- Artavanis-Tsakonas S., Rand, M.D., and Lake, R.J. 1999. Notch signaling: Cell fate control and signal integration in development. Science 284: 770–776. - PubMed
- Aybar M.J., Nieto, M.A., and Mayor, R. 2003. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130: 483–494. - PubMed
- Barrantes I.B., Elia, A.J., Wunsch, K., De Angelis, M.H., Mak, T.W., Rossant, J., Conlon, R.A., Gossler, A., and de la Pompa, J.L. 1999. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9: 470–480. - PubMed
- Batlle E., Sancho, E., Franci, C., Dominguez, D., Monfar, M., Baulida, J., and Garcia De Herreros, A. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2: 84–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials