A Method to Identify p62's UBA Domain Interacting Proteins - PubMed (original) (raw)
A Method to Identify p62's UBA Domain Interacting Proteins
Julia W. Pridgeon et al. Biol Proced Online. 2003.
Abstract
The UBA domain is a conserved sequence motif among polyubiquitin binding proteins. For the first time, we demonstrate a systematic, high throughput approach to identification of UBA domain-interacting proteins from a proteome-wide perspective. Using the rabbit reticulocyte lysate in vitro expression cloning system, we have successfully identified eleven proteins that interact with p62's UBA domain, and the majority of the eleven proteins are associated with neurodegenerative disorders, such as Alzheimer's disease. Therefore, p62 may play a novel regulatory role through its UBA domain. Our approach provides an easy route to the characterization of UBA domain interacting proteins and its application will unfold the important roles that the UBA domain plays.
Figures
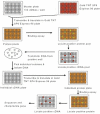
Fig. 1. Schematic description of the in vitro expression cloning (IVEC) system for the primary proteome-wide screen and subsequent isolation of cDNAs encoding the proteins of interest (from IVEC manual).
Fig. 2. Protein synthesized by the ProteoLink IVEC system in the presence of [35S] methionine.
96 protein pools were generated by employing TNT Quick Coupled in vitro transcription/translation system. The 96 protein pools were then divided into 24 mixed protein pools by combining four protein pools as one mixed protein pool. A: 1 day exposure; B: 3 days exposure.
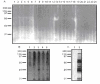
Fig. 3. Western blot analysis of the proteins synthesized by the ProteoLink IVEC system with ubiquitin antibody and specificity of UBA binding.
A: Mixed protein pools. 96 protein pools were generated by employing TNT Quick Coupled in vitro transcription/translation system. The 96 protein pools were divided into 24 mixed protein pools and separated by SDS-PAGE, followed by immunoblotting with ubiquitin monoclonal antibody. B: Proteins synthesized in the IVEC system in the absence of ubiquitin (lane 1) or presence of wild type ubiquitin (lane 2), ubiquitin K29 (lane 3), ubiquitin K48 (lane 4), and ubiquitin K63 (lane 5). C: Proteins synthesized in the IVEC system were labeled by 35S methionine and used for pull down assays. Lane 1: proteins recovered with agarose beads alone; Lane 2: proteins recovered with p62 UBA agarose beads.
Fig. 4. Pull down assay of p62’s UBA domain with proteins (mixed protein pool) synthesized by IVEC system in the presence of [35S] methionine.
The 24 mixed proteins pools were incubated with agarose-immobilised p62 UBA beads for 2 hours at 4ºC and washed three times in washing buffer. Bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. Positive “hits” were marked by arrows and the positive protein pools were selected for subsequent deconvulation.
Fig. 5. Pull down assay of p62’s UBA domain with proteins (individual protein pool) synthesized by IVEC system in the presence of [35S] methionine.
The 6 positive mix protein pools (# 2, 4, 8, 14, 20, 21 from Fig. 4) representing 24 individual protein pools were incubated with agarose-immobilised p62 UBA beads for 2 hours at 4ºC and the beads were washed three times in washing buffer. Bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. Positive “hits” were marked by arrows and the positive individual protein pools were underlined.
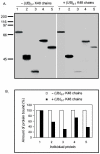
Fig. 6. Polyubiquitin K48 chain (Ub2-7 K48) competition pull down assay.
A: The individual proteins were synthesized by employing TNT Quick Coupled in vitro transcription/translation system and incubated with 5 mg agarose-immobilised p62 UBA beads +/- polyubiquitin K48 chains for 2 hours at 4ºC. The beads were washed three times in washing buffer and bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. B: The autoradiogram was scanned and the relative amount of protein (%) bound to p62’s UBA domain +/- polyubiquitin K48 chains was graphed. The amount of protein bound to p62’s UBA domain without addition of polyubiquitin K48 chains was considered 100%. 1: HSP70; 2: Meis2; 3: 14-3-3; 4: Reelin; 5: MBP.
Fig. 7. In vitro ubiquitination using the rabbit reticulocyte lysate.
A: HSP70 Protein was synthesized employing TNT Quick Coupled in vitro transcription/translation system in the presence of ubiquitin, resolved on 10% SDS-PAGE gels, transferred to nitrocellulose membrane and western blotted with ubiquitin monoclonal antibody. B: HSP70 Protein was synthesized employing TNT Quick Coupled in vitro transcription/translation system in the presence of ubiquitin and 35S-methionine, resolved on 10% SDS-PAGE and exposed to X-ray film. C: Western blot of rabbit reticulocyte lysate with TRAF6 (E3) and UbcH7 (E2).
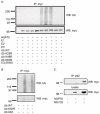
Fig. 8. In vitro ubiquitination of HSP70 using E1-E2-E3 system and in vivo interaction of p62 with HSP70.
A: myc-tagged HSP70 protein expressed in HEK cells was immunoprecipitated by myc-polyclonal antibody and used as a source of substrate for in vitro ubiquitination +/- E1, E2, E3, WT-Ub, Ub K29R, Ub K48R, Ub K63R. The reactions were separated on 10% SDS-PAGE and Western blotted with myc monoclonal antibody (bottom panel) to detect HSP70 or with ubiquitin monoclonal antibody (top panel) to detect ubiquitination. B: myc-tagged HSP70 protein expressed in HEK cells was immunoprecipitated by myc-polyclonal antibody and used as a source of substrate for in vitro ubiquitination in the absence or presence of wild type ubiquitin or Ub K63R or Ub K63. The reactions were separated on 10% SDS-PAGE and Western blotted with myc monoclonal antibody (bottom panel) to detect HSP70 or with ubiquitin monoclonal antibody (top panel) to detect ubiquitination. C: In vivo interaction of HSP70 and p62. Transfection of myc-tagged HSP70 into HEK293 cells was performed and the cell lysates were subjected to immunoprecipitation with p62 polyclonal antibody, followed by Western blot with anti-myc monoclonal antibody.
Similar articles
- Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62's role in neurodegenerative disease.
Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC. Wooten MW, et al. J Biomed Biotechnol. 2006;2006(3):62079. doi: 10.1155/JBB/2006/62079. J Biomed Biotechnol. 2006. PMID: 17047309 Free PMC article. - Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain.
Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Trempe JF, et al. EMBO J. 2005 Sep 21;24(18):3178-89. doi: 10.1038/sj.emboj.7600797. Epub 2005 Sep 1. EMBO J. 2005. PMID: 16138082 Free PMC article. - Structural and functional studies of mutations affecting the UBA domain of SQSTM1 (p62) which cause Paget's disease of bone.
Layfield R, Ciani B, Ralston SH, Hocking LJ, Sheppard PW, Searle MS, Cavey JR. Layfield R, et al. Biochem Soc Trans. 2004 Nov;32(Pt 5):728-30. doi: 10.1042/BST0320728. Biochem Soc Trans. 2004. PMID: 15493999 Review. - Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A.
Raasi S, Orlov I, Fleming KG, Pickart CM. Raasi S, et al. J Mol Biol. 2004 Aug 27;341(5):1367-79. doi: 10.1016/j.jmb.2004.06.057. J Mol Biol. 2004. PMID: 15321727 - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
- Analyzing microarray data of Alzheimer's using cluster analysis to identify the biomarker genes.
Guttula SV, Allam A, Gumpeny RS. Guttula SV, et al. Int J Alzheimers Dis. 2012;2012:649456. doi: 10.1155/2012/649456. Epub 2012 Feb 14. Int J Alzheimers Dis. 2012. PMID: 22482075 Free PMC article. - A small molecule inhibitor of inducible heat shock protein 70.
Leu JI, Pimkina J, Frank A, Murphy ME, George DL. Leu JI, et al. Mol Cell. 2009 Oct 9;36(1):15-27. doi: 10.1016/j.molcel.2009.09.023. Mol Cell. 2009. PMID: 19818706 Free PMC article. - Mutant p62P392L stimulation of osteoclast differentiation in Paget's disease of bone.
Sundaram K, Shanmugarajan S, Rao DS, Reddy SV. Sundaram K, et al. Endocrinology. 2011 Nov;152(11):4180-9. doi: 10.1210/en.2011-1225. Epub 2011 Aug 30. Endocrinology. 2011. PMID: 21878516 Free PMC article. - Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6.
Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Olzmann JA, et al. J Cell Biol. 2007 Sep 10;178(6):1025-38. doi: 10.1083/jcb.200611128. J Cell Biol. 2007. PMID: 17846173 Free PMC article. - Defining an Embedded Code for Protein Ubiquitination.
Jadhav T, Wooten MW. Jadhav T, et al. J Proteomics Bioinform. 2009 Jul 24;2:316. doi: 10.4172/jpb.1000091. J Proteomics Bioinform. 2009. PMID: 20148194 Free PMC article.
References
- Cooper JA, Howell B. The when and how of Src regulation. Cell. 1993;73:1051–1054. - PubMed
- Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources