Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress - PubMed (original) (raw)
Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress
Orit Uziel et al. J Bacteriol. 2004 Jan.
Abstract
In this report we describe the cloning, organization, and promoter analysis of the Staphylococcus aureus thioredoxin (trxA) and thioredoxin reductase (trxB) genes and their transcription in response to changes in oxygen concentration and to oxidative stress compounds. Northern analysis showed that the S. aureus trxA and trxB genes were transcribed equally well in aerobic and anaerobic conditions. Several oxidative stress compounds were found to rapidly induce transcription of the trxA and trxB genes. The most pronounced effects were seen with diamide, a thiol-specific oxidant that promotes disulfide bond formation; menadione, a redox cycling agent; and tau-butyl hydroperoxide, an organic peroxide. In each case the induction was independent of the general stress sigma factor sigma(B). These studies show that the S. aureus trxA and trxB genes are upregulated following exposure to these oxidative stress agents, resulting in increased disulfide bond formation. In contrast, no effect of hydrogen peroxide on induction of the trxA and trxB genes was seen. We also show that the S. aureus thioredoxin reductase appears to be essential for growth. This observation, coupled with structural differences between the bacterial and mammalian thioredoxin reductases, suggests that it may serve as a target for the development of new antimicrobials.
Figures
FIG. 1.
Chromosomal organization, primer extension analysis, and sequence of the S. aureus Oxford trxA gene. (A) The gene organization is identical to that found in all of the S. aureus strains referred to in this work; the numbering of nucleotides in intergenic regions is for S. aureus Mu50, accession no.. NC_002758. The ≈5.3-kbp DNA region containing trxA and flanking genes is shown. Gene designations: mutS2 encodes a mismatch repair ATPase; trxA encodes thioredoxin; uvrC encodes a subunit of the endonuclease nucleotide excision repair system. (B) Total RNA was isolated from aerobic cultures of S. aureus Oxford. Primer extension was carried out as described in Materials and Methods, and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primers. Arrows point to the A nucleotides of the two trxA transcription start sites. (C) Nucleotide sequence of the trxA promoter (numbering according to GenBank AJ223480). Transcription start points are shown by bent arrows above the underlined boldface A nucleotides. The trxA ATG translational start codon (underlined) and its ribosome-binding site are shown in boldface italic letters. Putative −10 and −35 hexamer sequences are shown as boxed boldface italic letters. Two direct repeats are shown by grey arrows above the nucleotide sequences. The TAA stop codons of the mutS2 and trxA genes are marked by an asterisk. A putative rho-independent transcriptional terminator is indicated by two solid inverted arrows below the nucleotide sequence.
FIG. 2.
Chromosomal organization, primer extension analysis, and sequence of the S. aureus Oxford trxB gene. (A) The organization is identical to that found in all of the S. aureus strains referred to in this work; the numbering of nucleotides in intergenic regions is for S. aureus Mu50, accession no. NC_002758. The ≈11.4-kbp DNA region containing trxB and flanking genes is shown. Gene designations: uvrA and uvrB encode subunits of the endonuclease nucleotide excision repair system; hprK encodes a bifunctional kinase-phosphatase_; lgt_ encodes a prelipoprotein diacylglyceroltransferase; yvoF encodes a putative acetyltransferase; yvcD encodes a hypothetical protein (designations according to reference 8); trxB encodes thioredoxin reductase. The directions of ORFs are indicated by arrows. Boxes indicate two intergenic regions that contain STAR tandem direct repeats. (B) Total RNA was isolated from aerobic cultures of S. aureus Oxford. Primer extension was carried as described in Materials and Methods, and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primers. An arrow points to the A nucleotide of the trxB transcription start point. (C) Nucleotide sequence of the trxB promoter (numbering according to GenBank AJ223781). The transcription start point is shown by a bent arrow above the underlined boldface A nucleotide. The trxB ATG translational start codon (underlined) and its ribosome-binding site are shown in boldface italic letters. Putative −10 and −35 hexamer sequences are shown as boxed boldface italic letters. The TAG and TAA translational stop codons of the yvcD and trxB genes, respectively, are marked by asterisks. Four pairs of inverted repeats located downstream of trxB are shown by arrows below the nucleotide sequence. Three long imperfect STAR direct repeats containing an _Apa_I endonuclease restriction site (GGGCCC) are indicated in bold letters.
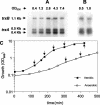
FIG. 3.
Northern hybridization analysis of S. aureus Oxford trxA and trxB transcripts in aerobic and anaerobic cultures. Total RNA was obtained from aerobic (A) and anaerobic (B) cultures at the early and mid-exponential phases and stationary phase, electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Sizes of transcripts are in kilobases; arrows indicate the positions of transcripts. (C) Growth profiles of S. aureus Oxford in aerobic and anaerobic conditions. The OD600 was used to follow the growth of cultures in TSB medium at 37°C. The arrows in panel C indicate the samples in panels A and B.
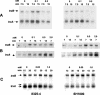
FIG. 4.
Northern hybridization analysis of S. aureus 8325-4 and SH1000 trxA and trxB transcripts: effect of diamide. Cultures of 8325-4 and SH1000 were grown aerobically to the mid-exponential phase of growth and treated with 0.5 and 2.0 mM diamide for 15 min. Total RNA obtained from cultures at an OD600 of ≈1.0 was electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Arrows indicate the positions of transcripts.
FIG. 5.
Northern hybridization analysis of S. aureus 8325-4 and SH1000 trxA and trxB transcripts: effect of oxidative stress agents. Cultures of 8325-4 and SH1000 were grown aerobically to the mid-exponential phase of growth and treated with (A) menadione, (B) τ-butyl hydroperoxide, and (C) hydrogen peroxide at the concentrations and for the times shown. Total RNA obtained from cultures at an OD600 of ≈1.0 was electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Arrows indicate the positions of transcripts.
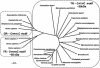
FIG. 6.
Phylogeny of bacterial and human thioredoxin and glutathione reductases (TR and GR, respectively). The deduced amino acid sequences of thioredoxin reductase were determined for the proteins from Brucella suis (GenBank AE014445), Chlamydia pneumoniae (SwissProt Q9Z8M4), Clostridium tetani (SwissProt Q890T3), Coxiella burnetii (GenBank X75627), Escherichia coli (SwissProt P09625), Haemophilus influenzae (SwissProt P43788), Helicobacter pylori (SwissProt P56431), Homo sapiens 1 (SwissProt Q16881), H. sapiens 2 (GenBank AF171055), H. sapiens 3 (GenBank AF171054), Listeria monocytogenes (SwissProt O32823), Mycobacterium leprae (SwissProt P46843), Mycobacterium tuberculosis (SwissProt P52214), Mycoplasma genitalium (SwissProt P47348), Neisseria meningitides (GenBank AL162756), Pseudomonas aeruginosa (SwissProt Q9I0M2), Rickettsia prowazekii (SwissProt Q9ZD33), Staphylococcus aureus (SwissProt O54079), Streptococcus pyogenes (SwissProt Q878I8), Treponema pallidum (SwissProt O83790), Vibrio cholerae (SwissProt Q9KSS4), and Yersinia pestis (Swiss Prot: Q8ZGC9). Deduced amino acid sequences of glutathione reductases used were from Escherichia coli (SwissProt P06715), Haemophilus influenzae (SwissProt P43783), Homo sapiens (SwissProt P00390), and Streptococcus pyogenes (GenBank AE014147).
Similar articles
- Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis.
Rocha ER, Tzianabos AO, Smith CJ. Rocha ER, et al. J Bacteriol. 2007 Nov;189(22):8015-23. doi: 10.1128/JB.00714-07. Epub 2007 Sep 14. J Bacteriol. 2007. PMID: 17873045 Free PMC article. - Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei.
Serata M, Iino T, Yasuda E, Sako T. Serata M, et al. Microbiology (Reading). 2012 Apr;158(Pt 4):953-962. doi: 10.1099/mic.0.053942-0. Epub 2012 Feb 2. Microbiology (Reading). 2012. PMID: 22301908 - Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus.
Ballal A, Manna AC. Ballal A, et al. J Bacteriol. 2010 Jan;192(1):336-45. doi: 10.1128/JB.01202-09. J Bacteriol. 2010. PMID: 19854896 Free PMC article. - The thioredoxin antioxidant system.
Lu J, Holmgren A. Lu J, et al. Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review. - Physiological functions of thioredoxin and thioredoxin reductase.
Arnér ES, Holmgren A. Arnér ES, et al. Eur J Biochem. 2000 Oct;267(20):6102-9. doi: 10.1046/j.1432-1327.2000.01701.x. Eur J Biochem. 2000. PMID: 11012661 Review.
Cited by
- Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response.
Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. Painter KL, et al. Infect Immun. 2015 May;83(5):1830-44. doi: 10.1128/IAI.03016-14. Epub 2015 Feb 17. Infect Immun. 2015. PMID: 25690100 Free PMC article. - Physiological and expression analyses of Agrobacterium tumefaciens trxA, encoding thioredoxin.
Vattanaviboon P, Tanboon W, Mongkolsuk S. Vattanaviboon P, et al. J Bacteriol. 2007 Sep;189(17):6477-81. doi: 10.1128/JB.00623-07. Epub 2007 Jun 15. J Bacteriol. 2007. PMID: 17573482 Free PMC article. - Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes.
Zeller T, Klug G. Zeller T, et al. Naturwissenschaften. 2006 Jun;93(6):259-66. doi: 10.1007/s00114-006-0106-1. Naturwissenschaften. 2006. PMID: 16555095 Review. - Steady-state hydrogen peroxide induces glycolysis in Staphylococcus aureus and Pseudomonas aeruginosa.
Deng X, Liang H, Ulanovskaya OA, Ji Q, Zhou T, Sun F, Lu Z, Hutchison AL, Lan L, Wu M, Cravatt BF, He C. Deng X, et al. J Bacteriol. 2014 Jul;196(14):2499-513. doi: 10.1128/JB.01538-14. Epub 2014 Apr 25. J Bacteriol. 2014. PMID: 24769698 Free PMC article.
References
- Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. - PubMed
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources