A map of the interactome network of the metazoan C. elegans - PubMed (original) (raw)
. 2004 Jan 23;303(5657):540-3.
doi: 10.1126/science.1091403. Epub 2004 Jan 2.
Christopher M Armstrong, Nicolas Bertin, Hui Ge, Stuart Milstein, Mike Boxem, Pierre-Olivier Vidalain, Jing-Dong J Han, Alban Chesneau, Tong Hao, Debra S Goldberg, Ning Li, Monica Martinez, Jean-François Rual, Philippe Lamesch, Lai Xu, Muneesh Tewari, Sharyl L Wong, Lan V Zhang, Gabriel F Berriz, Laurent Jacotot, Philippe Vaglio, Jérôme Reboul, Tomoko Hirozane-Kishikawa, Qianru Li, Harrison W Gabel, Ahmed Elewa, Bridget Baumgartner, Debra J Rose, Haiyuan Yu, Stephanie Bosak, Reynaldo Sequerra, Andrew Fraser, Susan E Mango, William M Saxton, Susan Strome, Sander Van Den Heuvel, Fabio Piano, Jean Vandenhaute, Claude Sardet, Mark Gerstein, Lynn Doucette-Stamm, Kristin C Gunsalus, J Wade Harper, Michael E Cusick, Frederick P Roth, David E Hill, Marc Vidal
Affiliations
- PMID: 14704431
- PMCID: PMC1698949
- DOI: 10.1126/science.1091403
A map of the interactome network of the metazoan C. elegans
Siming Li et al. Science. 2004.
Abstract
To initiate studies on how protein-protein interaction (or "interactome") networks relate to multicellular functions, we have mapped a large fraction of the Caenorhabditis elegans interactome network. Starting with a subset of metazoan-specific proteins, more than 4000 interactions were identified from high-throughput, yeast two-hybrid (HT=Y2H) screens. Independent coaffinity purification assays experimentally validated the overall quality of this Y2H data set. Together with already described Y2H interactions and interologs predicted in silico, the current version of the Worm Interactome (WI5) map contains approximately 5500 interactions. Topological and biological features of this interactome network, as well as its integration with phenome and transcriptome data sets, lead to numerous biological hypotheses.
Figures
Fig. 1
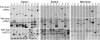
Coaffinity purification assays. Shown are 10 examples from the Core-1, Core-2, and Non-Core data sets. The top panels show Myc-tagged prey expression after affinity purification on glutathione-Sepharose, demonstrating binding to GST-bait. The middle and bottom panels show expression of Myc-prey and GST-bait, respectively. The lanes alternate between extracts expressing GST-bait proteins (+) and GST alone (-). ORF pairs are identified in table S1 with the lane number corresponding to the order in which they appear in the table.
Fig.2
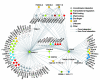
Analysis of the WI5 network. (A) Nodes (representing proteins) are colored according to their phylogenic class: ancient (red), multicellular (yellow), and worm (blue). Edges represent protein-protein interactions. The inset highlights a small part of the network. (B) The proportion of proteins, P(k), with different numbers of interacting partners, k, is shown for C. elegans proteins used as baits or preys and for S. cerevisiae proteins. (C) The pie charts show the proportion of interacting preys found in Y2H screens that fall into each phylogenic class. Also shown is the distribution of all preys found and all preys searched in the AD-ORFeome1.0 library. (D) Overlap with transcriptome (see text) (18), Pearson correlation coefficients (PCCs) were calculated and graphed for each pair of proteins in the interaction data sets and their corresponding randomized data sets. The red area to the right corresponds to interactions that show a significant relationship to expression profiling data (P < 0.05). (E) Interactions between proteins in Topomap mountain 29 (18). The dash-circled proteins belong to the same paralogous family (sharing more than 80% homology) and are thus collapsed into one set of interactions. (F) Proportion of interaction pairs where both genes are embryonic lethal (P < 10-7).
Fig.3
Graphical representation of a highly interconnected subnetwork around VAB-3 and C49A1.4. Biological functional classes were obtained from WormPD (10).
Similar articles
- Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network.
Tewari M, Hu PJ, Ahn JS, Ayivi-Guedehoussou N, Vidalain PO, Li S, Milstein S, Armstrong CM, Boxem M, Butler MD, Busiguina S, Rual JF, Ibarrola N, Chaklos ST, Bertin N, Vaglio P, Edgley ML, King KV, Albert PS, Vandenhaute J, Pandey A, Riddle DL, Ruvkun G, Vidal M. Tewari M, et al. Mol Cell. 2004 Feb 27;13(4):469-82. doi: 10.1016/s1097-2765(04)00033-4. Mol Cell. 2004. PMID: 14992718 - Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline.
Walhout AJ, Reboul J, Shtanko O, Bertin N, Vaglio P, Ge H, Lee H, Doucette-Stamm L, Gunsalus KC, Schetter AJ, Morton DG, Kemphues KJ, Reinke V, Kim SK, Piano F, Vidal M. Walhout AJ, et al. Curr Biol. 2002 Nov 19;12(22):1952-8. doi: 10.1016/s0960-9822(02)01279-4. Curr Biol. 2002. PMID: 12445390 - Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network.
Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi-Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. Simonis N, et al. Nat Methods. 2009 Jan;6(1):47-54. doi: 10.1038/nmeth.1279. Nat Methods. 2009. PMID: 19123269 Free PMC article. - Networks in Caenorhabditis elegans.
Gunsalus KC, Rhrissorrakrai K. Gunsalus KC, et al. Curr Opin Genet Dev. 2011 Dec;21(6):787-98. doi: 10.1016/j.gde.2011.10.003. Epub 2011 Nov 4. Curr Opin Genet Dev. 2011. PMID: 22054717 Review. - Mapping the Protein-Protein Interactome Networks Using Yeast Two-Hybrid Screens.
Rajagopala SV. Rajagopala SV. Adv Exp Med Biol. 2015;883:187-214. doi: 10.1007/978-3-319-23603-2_11. Adv Exp Med Biol. 2015. PMID: 26621469 Free PMC article. Review.
Cited by
- A field-proven yeast two-hybrid protocol used to identify coronavirus-host protein-protein interactions.
Vidalain PO, Jacob Y, Hagemeijer MC, Jones LM, Neveu G, Roussarie JP, Rottier PJ, Tangy F, de Haan CA. Vidalain PO, et al. Methods Mol Biol. 2015;1282:213-29. doi: 10.1007/978-1-4939-2438-7_18. Methods Mol Biol. 2015. PMID: 25720483 Free PMC article. - FR database 1.0: a resource focused on fruit development and ripening.
Yue J, Ma X, Ban R, Huang Q, Wang W, Liu J, Liu Y. Yue J, et al. Database (Oxford). 2015 Feb 27;2015:bav002. doi: 10.1093/database/bav002. Print 2015. Database (Oxford). 2015. PMID: 25725058 Free PMC article. - Kinesin-1 prevents capture of the oocyte meiotic spindle by the sperm aster.
McNally KL, Fabritius AS, Ellefson ML, Flynn JR, Milan JA, McNally FJ. McNally KL, et al. Dev Cell. 2012 Apr 17;22(4):788-98. doi: 10.1016/j.devcel.2012.01.010. Epub 2012 Mar 29. Dev Cell. 2012. PMID: 22465668 Free PMC article. - Gene-centered regulatory network mapping.
Walhout AJ. Walhout AJ. Methods Cell Biol. 2011;106:271-88. doi: 10.1016/B978-0-12-544172-8.00010-4. Methods Cell Biol. 2011. PMID: 22118281 Free PMC article. Review. - The ubiquitin-fold modifier 1 (Ufm1) cascade of Caenorhabditis elegans.
Hertel P, Daniel J, Stegehake D, Vaupel H, Kailayangiri S, Gruel C, Woltersdorf C, Liebau E. Hertel P, et al. J Biol Chem. 2013 Apr 12;288(15):10661-71. doi: 10.1074/jbc.M113.458000. Epub 2013 Feb 28. J Biol Chem. 2013. PMID: 23449979 Free PMC article.
References
- Marcotte EM, et al. Science. 1999;285:751. - PubMed
- Uetz P, et al. Nature. 2000;403:623. - PubMed
- Ho Y, et al. Nature. 2002;415:180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases