A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization - PubMed (original) (raw)
Comparative Study
doi: 10.1101/gr.1878804.
Jos Jonkers, Hannah Kitson, Heike Fiegler, Sean Humphray, Carol Scott, Sarah Hunt, Yuejin Yu, Ichiko Nishijima, Arno Velds, Henne Holstege, Nigel Carter, Allan Bradley
Affiliations
- PMID: 14707179
- PMCID: PMC314296
- DOI: 10.1101/gr.1878804
Comparative Study
A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization
Yeun-Jun Chung et al. Genome Res. 2004 Jan.
Abstract
Microarray-based comparative genomic hybridization (CGH) has become a powerful method for the genome-wide detection of chromosomal imbalances. Although BAC microarrays have been used for mouse CGH studies, the resolving power of these analyses was limited because high-density whole-genome mouse BAC microarrays were not available. We therefore developed a mouse BAC microarray containing 2803 unique BAC clones from mouse genomic libraries at 1-Mb intervals. For the general amplification of BAC clone DNA prior to spotting, we designed a set of three novel degenerate oligonucleotide-primed (DOP) PCR primers that preferentially amplify mouse genomic sequences while minimizing unwanted amplification of contaminating Escherichia coli DNA. The resulting 3K mouse BAC microarrays reproducibly identified DNA copy number alterations in cell lines and primary tumors, such as single-copy deletions, regional amplifications, and aneuploidy.
Figures
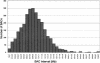
Figure 1
Distribution of spacing of the 2825 BACs used for the 3K mouse BAC microarray. The average spacing of the BACs was 0.89 Mb across the 19 autosomes and chromosome X. Linear map positions and other details of the 3K mouse BAC clone set are available online from the Ensembl database (http://www.ensembl.org/Mus\_musculus/cytoview).
Figure 2
Detection of single-copy differences. (A) Normal vs. normal hybridization of spleen DNA from a C57BL/6J female. (B) Comparative genomic hybridization of normal C57BL/6J female DNA vs. normal C57BL/6J male DNA. Plotted are log2-transformed hybridization ratios against the linear map position of the BACs (in Mb). Confidence levels were calculated according to the Rosetta Error Model. Red indicates significant gain in DNA copy number, green indicates significant loss, and black indicates no significant change. The CGH profile of the female vs. male hybridization shows identical DNA copy numbers for all autosomes (mean log2 ratio -0.0066, s.d. of the log2 ratios = 0.064); the copy numbers of male and female chromosome X sequences were clearly different (mean log2 ratio of 0.63).
Figure 3
Detection of regional single-copy deletions. (A) Comparative genomic hybridization of normal AB2.2 ES cell DNA vs. DNA from 7A9 ES cells, harboring a regional single-copy deletion between D4Mit117 (94.6 Mb) and D4Mit246 (96.6 Mb) on chromosome 4. (B) Comparative genomic hybridization of normal AB2.2 ES cell DNA vs. DNA from 3D5 ES cells, harboring a 6.9-Mb single-copy deletion from Mpo2 (88.5 Mb) to Chad2 (95.4 Mb) on chromosome 11. The BAC clones spanning the deletion regions (green dots) are denoted. Both deletions were confirmed by FISH analysis. On prometaphase chromosomes from 7A9 and 3D5 ES cells, only one hybridization signal was observed, with probes corresponding to the deleted region (red signals, indicated by red arrow), whereas two hybridization signals, derived from both alleles, were detected with probes located outside of the deletion regions (green signals, indicated by green arrows).
Figure 4
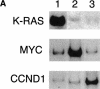
Array CGH analysis of oncogene amplifications in mouse mammary tumors. (A) Southern analysis of tumor DNA using Kras2, Myc, and Ccnd1 specific probes. Tumors 1, 2, and 3 showed amplification of Kras2, Myc, and Ccnd1, respectively. (B) Whole-genome array CGH profiles (left panels) and CGH profiles of the relevant chromosomes (right panels) from mouse mammary tumors 1, 2, and 3. Plotted are log2-transformed hybridization ratios of tumor DNA vs. control DNA. Vertical red bars on each chromosome indicate the positions of Kras2, Myc, and Ccnd1, respectively.
Figure 4
Array CGH analysis of oncogene amplifications in mouse mammary tumors. (A) Southern analysis of tumor DNA using Kras2, Myc, and Ccnd1 specific probes. Tumors 1, 2, and 3 showed amplification of Kras2, Myc, and Ccnd1, respectively. (B) Whole-genome array CGH profiles (left panels) and CGH profiles of the relevant chromosomes (right panels) from mouse mammary tumors 1, 2, and 3. Plotted are log2-transformed hybridization ratios of tumor DNA vs. control DNA. Vertical red bars on each chromosome indicate the positions of Kras2, Myc, and Ccnd1, respectively.
Figure 5
Array CGH analysis of in vitro transformed RMT18 MEFs derived from a female embryo. (A) CGH profiles of all autosomes and the X-chromosome, showing copy number gains for chromosomes 6, 10, and 19, and copy number loss for chromosome X. (B) Ploidy analysis of RMT18 cells. Normal mouse ES cells were used as a normal diploid control. DNA index is 1.37. (_C_-E) Chromosome painting of RMT18 metaphase spreads for chromosome 19, X, and 18, respectively.
Similar articles
- DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones.
Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP. Fiegler H, et al. Genes Chromosomes Cancer. 2003 Apr;36(4):361-74. doi: 10.1002/gcc.10155. Genes Chromosomes Cancer. 2003. PMID: 12619160 - Methods for high throughput validation of amplified fragment pools of BAC DNA for constructing high resolution CGH arrays.
Watson SK, deLeeuw RJ, Ishkanian AS, Malloff CA, Lam WL. Watson SK, et al. BMC Genomics. 2004 Jan 14;5(1):6. doi: 10.1186/1471-2164-5-6. BMC Genomics. 2004. PMID: 14723794 Free PMC article. - Method for manufacturing whole-genome microarrays by rolling circle amplification.
Smirnov DA, Burdick JT, Morley M, Cheung VG. Smirnov DA, et al. Genes Chromosomes Cancer. 2004 May;40(1):72-7. doi: 10.1002/gcc.20015. Genes Chromosomes Cancer. 2004. PMID: 15034872 - MAPH: from gels to microarrays.
Patsalis PC, Kousoulidou L, Sismani C, Männik K, Kurg A. Patsalis PC, et al. Eur J Med Genet. 2005 Jul-Sep;48(3):241-9. doi: 10.1016/j.ejmg.2005.04.011. Eur J Med Genet. 2005. PMID: 16179220 Review. - [Array-CGH for routine diagnosis of cryptic chromosomal imbalances].
Andrieux J. Andrieux J. Pathol Biol (Paris). 2008 Sep;56(6):368-74. doi: 10.1016/j.patbio.2008.04.011. Epub 2008 Jun 2. Pathol Biol (Paris). 2008. PMID: 18514435 Review. French.
Cited by
- Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling.
van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. van Harn T, et al. Genes Dev. 2010 Jul 1;24(13):1377-88. doi: 10.1101/gad.580710. Epub 2010 Jun 15. Genes Dev. 2010. PMID: 20551164 Free PMC article. - Candidate metastasis suppressor genes uncovered by array comparative genomic hybridization in a mouse allograft model of prostate cancer.
Yi Y, Nandana S, Case T, Nelson C, Radmilovic T, Matusik RJ, Tsuchiya KD. Yi Y, et al. Mol Cytogenet. 2009 Sep 26;2:18. doi: 10.1186/1755-8166-2-18. Mol Cytogenet. 2009. PMID: 19781100 Free PMC article. - Loss of Rassf1a cooperates with Apc(Min) to accelerate intestinal tumourigenesis.
van der Weyden L, Arends MJ, Dovey OM, Harrison HL, Lefebvre G, Conte N, Gergely FV, Bradley A, Adams DJ. van der Weyden L, et al. Oncogene. 2008 Jul 24;27(32):4503-8. doi: 10.1038/onc.2008.94. Epub 2008 Apr 7. Oncogene. 2008. PMID: 18391979 Free PMC article. - A genome assembly-integrated dog 1 Mb BAC microarray: a cytogenetic resource for canine cancer studies and comparative genomic analysis.
Thomas R, Duke SE, Karlsson EK, Evans A, Ellis P, Lindblad-Toh K, Langford CF, Breen M. Thomas R, et al. Cytogenet Genome Res. 2008;122(2):110-21. doi: 10.1159/000163088. Epub 2008 Dec 18. Cytogenet Genome Res. 2008. PMID: 19096206 Free PMC article. - Genetic alteration and gene expression modulation during cancer progression.
Garnis C, Buys TP, Lam WL. Garnis C, et al. Mol Cancer. 2004 Mar 22;3:9. doi: 10.1186/1476-4598-3-9. Mol Cancer. 2004. PMID: 15035667 Free PMC article. Review.
References
- Cai, W.W., Mao, J.H., Chow, C.W., Damani, S., Balmain, A., and Bradley, A. 2002. Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat. Biotech. 20: 393-396. - PubMed
- Carter, N.P., Ferguson-Smith, M.A., Perryman, M.T., Telenius, H., Pelmear, A.H., Leversha, M.A., Glancy, M.T., Wood, S.L., Cook, K., Dyson, H.M., et al. 1992. Reverse chromosome painting: A method for the rapid analysis of aberrant chromosomes in clinical cytogenetics. J. Med. Genet. 29: 299-307. - PMC - PubMed
- Darzynkiewicz, Z. and Juan, G. 1997. DNA content analysis of fixed cells with DAPI. In Current protocols in cytometry (ed. J.P. Robinson), pp. 7.5.3. J. Wiley, New York.
- Dietrich, W.F., Miller, J.C., Steen, R.G., Merchant, M., Damron, D., Nahf, R., Gross, A., Joyce, D.C., Wessel, M., Dredge, R.D., et al. 1994a. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat. Genet. 7: 220-245. - PubMed
WEB SITE REFERENCES
- http://www.ensembl.org/mus_musculus/cytoview; map positions of 1-Mb mouse BAC clone set.
- http://www.sanger.ac.uk/Software/Image; fingerprint gel image analysis software.
- http://www.bcgsc.ca/lab/mapping/mouse; mouse BAC fingerprints reference database.
- http://www.sanger.ac.uk/Projects/Microarrays/arraylab/protocol4; microarray spotting and post-spotting processing protocols.
- http://download.cell.com/supplementarydata/cell/102/1/109/DC1/ErModlv2.htm; Rosetta error model.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous