D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a - PubMed (original) (raw)
D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a
Graham Rena et al. EMBO Rep. 2004 Jan.
Abstract
The protein kinase CK1 phosphorylates serine residues that are located close to another phosphoserine in the consensus pSer-Xaa-Xaa-Ser. This specificity generates regions in its target proteins containing two or more neighbouring phosphoserine residues, termed here multisite phosphorylation domains (MPDs). In this paper, we demonstrate that D4476 is a potent and rather selective inhibitor of CK1 in vitro and in cells. In H4IIE hepatoma cells, D4476 specifically inhibits the phosphorylation of endogenous forkhead box transcription factor O1a (FOXO1a) on Ser322 and Ser325 within its MPD, without affecting the phosphorylation of other sites. Our results indicate that these residues are targeted by CK1 in vivo and that the CK1-mediated phosphorylation of the MPD is required for accelerated nuclear exclusion of FOXO1a in response to IGF-1 and insulin. D4476 is much more potent and specific than IC261 or CKI-7, and is therefore the most useful CK1 inhibitor currently available for identifying physiological substrates of CK1.
Figures
Figure 1
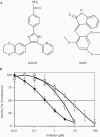
Structures of CK1 inhibitors and their effects on CK1 activity in vitro. (A) Structures of D4476 and IC261 (Mashhoon et al, 2000). (B) Purified CK1δ was assayed with phosphorylated peptide TFRPRTSpSNASTIS (30 μM), corresponding to the sequence surrounding residues 312–325 of FOXO1a, at an ATP concentration of 0.1 mM and varying concentrations of D4476 (filled circles), IC261 (open squares) and CKI-7 (open circles). The results are the average of at least four separate determinations, each performed in duplicate.
Figure 2
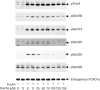
D4476 inhibits the phosphorylation of FOXO1a at Ser322 and Ser325, but not at Thr24 in vitro. Bacterially expressed GST-FOXO1a (1 μM) was left unphosphorylated (−) or maximally phosphorylated for 30 min at 30°C with 1 U ml−1 PKB, 10 mM magnesium acetate and 0.1 mM ATP, followed by phosphorylation for 10 min with 30 mU ml−1 CK1 (+), in the presence of the indicated concentrations of D4476. Aliquots of the reaction were spotted onto nitrocellulose and immunoblotted with phospho-specific antibodies recognizing phosphorylated Thr24 (pThr24), Ser322 (pSer322) and Ser325 (pSer325) and an antibody that recognizes the phosphorylated and unphosphorylated forms of FOXO1a equally well (FOXO1a). Similar results were obtained in several independent experiments.
Figure 3
D4476 specifically inhibits the phosphorylation at Ser322 and Ser325 specifically in H4IIE cells. Cells were serum starved for 4 h, and then stimulated for 30 min without (−) or with (+) 20 nM insulin. Aliquots of the cell lysates (2.5 mg protein) were then immunoprecipitated with 20 μg of anti-FOXO1a antibody raised against the whole protein. After washing the immunoprecipitates and denaturation in SDS, aliquots of the solubilized material were subjected to SDS–polyacrylamide gel electrophoresis. Following transfer to nitrocellulose, the membranes were subjected to immunoblotting using the antibodies used in Fig 1, as well as with a phospho-specific antibody that recognizes Ser329 (pSer329). Where indicated, the cells were incubated for 60 min with various concentrations of D4476 prior to stimulation with insulin. Similar results were obtained in at least six independent experiments.
Figure 4
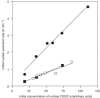
D4476 inhibits IGF-1 and serum-stimulated nuclear exclusion of FOXO1a in living cells. HEK293 cells were transfected with wild-type FOXO1a–GFP or mutant (Ser319Ala) FOXO1a–GFP. At 10 h post-transfection, the cells were serum starved for 12 h, and then stimulated in the presence of 50 ng ml−1 IGF-1 and 10% fetal calf serum with or without 150 μM D4476 pretreatment (10 min). Initial rates of nuclear exclusion were determined as described previously (Rena et al, 2002) by time-lapse imaging of live cells using confocal microscopy on a heated stage. Closed circles indicate the initial nuclear exclusion rate of FOXO1a in the absence of D4476. Open circles indicate the initial nuclear exclusion rate of FOXO1a in the presence of D4476. Squares indicate the initial nuclear exclusion rate of Ser319Ala FOXO1a–GFP. Each point is the average of seven cells.
Similar articles
- CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability.
Huart AS, MacLaine NJ, Meek DW, Hupp TR. Huart AS, et al. J Biol Chem. 2009 Nov 20;284(47):32384-94. doi: 10.1074/jbc.M109.052647. Epub 2009 Sep 15. J Biol Chem. 2009. PMID: 19759023 Free PMC article. - CK1 inhibitor affects in vitro maturation and developmental competence of bovine oocytes.
Shi P, Xu J, Zhao X, Shen P, Wen D, Yu Q, Deng Y, Shi D, Lu F. Shi P, et al. Reprod Domest Anim. 2019 Aug;54(8):1104-1112. doi: 10.1111/rda.13483. Epub 2019 Jun 23. Reprod Domest Anim. 2019. PMID: 31155763 - Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion.
Rena G, Woods YL, Prescott AR, Peggie M, Unterman TG, Williams MR, Cohen P. Rena G, et al. EMBO J. 2002 May 1;21(9):2263-71. doi: 10.1093/emboj/21.9.2263. EMBO J. 2002. PMID: 11980723 Free PMC article. - Achieving effective and selective CK1 inhibitors through structure modification.
Du C, Yang H, Feng F, Liu W, Chen Y, Sun H. Du C, et al. Future Med Chem. 2021 Mar;13(5):505-528. doi: 10.4155/fmc-2020-0215. Epub 2021 Jan 13. Future Med Chem. 2021. PMID: 33438471 Review. - Posttranslational regulation of Neurospora circadian clock by CK1a-dependent phosphorylation.
Querfurth C, Diernfellner A, Heise F, Lauinger L, Neiss A, Tataroglu O, Brunner M, Schafmeier T. Querfurth C, et al. Cold Spring Harb Symp Quant Biol. 2007;72:177-83. doi: 10.1101/sqb.2007.72.025. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419275 Review.
Cited by
- Casein kinase 1α inhibits p53 downstream of MDM2‑mediated autophagy and apoptosis in acute myeloid leukemia.
Xu W, Huang Z, Gan Y, Chen R, Huang Y, Xue B, Jiang S, Yu Z, Yu K, Zhang S. Xu W, et al. Oncol Rep. 2020 Nov;44(5):1895-1904. doi: 10.3892/or.2020.7760. Epub 2020 Sep 9. Oncol Rep. 2020. PMID: 32901886 Free PMC article. - Casein kinase I delta controls centrosome positioning during T cell activation.
Zyss D, Ebrahimi H, Gergely F. Zyss D, et al. J Cell Biol. 2011 Nov 28;195(5):781-97. doi: 10.1083/jcb.201106025. J Cell Biol. 2011. PMID: 22123863 Free PMC article. - F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress.
Malonia SK, Dutta P, Santra MK, Green MR. Malonia SK, et al. Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8632-7. doi: 10.1073/pnas.1510929112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124108 Free PMC article. - Casein Kinase 1α-A Target for Prostate Cancer Therapy?
Lishman-Walker E, Coffey K. Lishman-Walker E, et al. Cancers (Basel). 2024 Jul 2;16(13):2436. doi: 10.3390/cancers16132436. Cancers (Basel). 2024. PMID: 39001502 Free PMC article. Review. - 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1δ/ε.
Bischof J, Leban J, Zaja M, Grothey A, Radunsky B, Othersen O, Strobl S, Vitt D, Knippschild U. Bischof J, et al. Amino Acids. 2012 Oct;43(4):1577-91. doi: 10.1007/s00726-012-1234-x. Epub 2012 Feb 14. Amino Acids. 2012. PMID: 22331384 Free PMC article.
References
- Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR (1997) Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275: 1930–1933 - PubMed
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 96: 857–868 - PubMed
- Callahan JF et al. (2002) Identification of novel inhibitors of the transforming growth factor-1 (TGF-1) type 1 receptor (ALK5). J Med Chem 45: 999–1001 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous