Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection - PubMed (original) (raw)
Comparative Study
doi: 10.1086/381505. Epub 2004 Jan 7.
Affiliations
- PMID: 14712420
- PMCID: PMC1181921
- DOI: 10.1086/381505
Comparative Study
Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection
J L Elson et al. Am J Hum Genet. 2004 Feb.
Abstract
This article provides evidence that selection has been a significant force during the evolution of the human mitochondrial genome. Both gene-by-gene and whole-genome approaches were used here to assess selection in the 560 mitochondrial DNA (mtDNA) coding-region sequences that were used previously for reduced-median-network analysis. The results of the present analyses were complex, in that the action of selection was not indicated by all tests, but this is not surprising, in view of the characteristics and limitations of the different analytical methods. Despite these limitations, there is evidence for both gene-specific and lineage-specific variation in selection. Whole-genome sliding-window approaches indicated a lack of selection in large-scale segments of the coding region. In other tests, we analyzed the ratio of nonsynonymous-to-synonymous substitutions in the 13 protein-encoding mtDNA genes. The most straightforward interpretation of those results is that negative selection has acted on the mtDNA during evolution. Single-gene analyses indicated significant departures from neutrality in the CO1, ND4, and ND6 genes, although the data also suggested the possible operation of positive selection on the AT6 gene. Finally, our results and those of other investigators do not support a simple model in which climatic adaptation has been a major force during human mtDNA evolution.
Figures
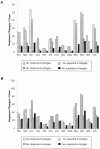
Figure 1
Plots of sequence changes/gene for haplogroup-associated (A) and private (B) polymorphisms. The plots contain the observed number of substitutions within each of the two major classes (the exact numbers are presented in table 1). The expected numbers of substitutions in each class are based on the total number of changes in that class and the predicted number of sites in each gene (determined using the DnaSP program [Rozas and Rozas 1999]). For the haplogroup-associated synonymous polymorphisms, a contingency test of the neutral expectation yielded a P value >.2 (12 df); thus, the data provide no evidence to reject the neutral model. These results indicate that there are no mutational hotspots for synonymous substitutions in the coding region. In contrast, when the nonsynonymous changes were analyzed, there was a clear deviation from the neutral expectation, and the contingency test yielded a P value <.001. For the private synonymous polymorphisms (_B_), a contingency test against the neutral expectation yielded a _P_ value >.05 (12 df); thus, the neutral model holds. In contrast, when the data for the nonsynonymous changes were analyzed, there was a clear deviation from the neutral expectation, and the contingency test yielded a P value <.001.
Similar articles
- Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J.
Moilanen JS, Finnila S, Majamaa K. Moilanen JS, et al. Mol Biol Evol. 2003 Dec;20(12):2132-42. doi: 10.1093/molbev/msg230. Epub 2003 Aug 29. Mol Biol Evol. 2003. PMID: 12949126 - Landscape genomics: natural selection drives the evolution of mitogenome in penguins.
Ramos B, González-Acuña D, Loyola DE, Johnson WE, Parker PG, Massaro M, Dantas GPM, Miranda MD, Vianna JA. Ramos B, et al. BMC Genomics. 2018 Jan 16;19(1):53. doi: 10.1186/s12864-017-4424-9. BMC Genomics. 2018. PMID: 29338715 Free PMC article. - Comparative genomics of mitochondrial DNA in Drosophila simulans.
Ballard JW. Ballard JW. J Mol Evol. 2000 Jul;51(1):64-75. doi: 10.1007/s002390010067. J Mol Evol. 2000. PMID: 10903373 - Positive and negative selection on the mitochondrial genome.
Meiklejohn CD, Montooth KL, Rand DM. Meiklejohn CD, et al. Trends Genet. 2007 Jun;23(6):259-63. doi: 10.1016/j.tig.2007.03.008. Epub 2007 Apr 5. Trends Genet. 2007. PMID: 17418445 Review. - Comparative genomics and the study of evolution by natural selection.
Ellegren H. Ellegren H. Mol Ecol. 2008 Nov;17(21):4586-96. doi: 10.1111/j.1365-294X.2008.03954.x. Mol Ecol. 2008. PMID: 19140982 Review.
Cited by
- Mitochondrial DNA sequence variation in Finnish patients with matrilineal diabetes mellitus.
Soini HK, Moilanen JS, Finnila S, Majamaa K. Soini HK, et al. BMC Res Notes. 2012 Jul 10;5:350. doi: 10.1186/1756-0500-5-350. BMC Res Notes. 2012. PMID: 22780954 Free PMC article. - Can parallel mutation and neutral genome selection explain Eastern African M1 consensus HVS-I motifs in Indian M haplogroups.
Winters C. Winters C. Indian J Hum Genet. 2007 Sep;13(3):93-6. doi: 10.4103/0971-6866.38982. Indian J Hum Genet. 2007. PMID: 21957355 Free PMC article. No abstract available. - Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations.
Greaves LC, Elson JL, Nooteboom M, Grady JP, Taylor GA, Taylor RW, Mathers JC, Kirkwood TB, Turnbull DM. Greaves LC, et al. PLoS Genet. 2012;8(11):e1003082. doi: 10.1371/journal.pgen.1003082. Epub 2012 Nov 15. PLoS Genet. 2012. PMID: 23166522 Free PMC article. - Comparative analysis of mitochondrial genomes in Bombina (Anura; Bombinatoridae).
Pabijan M, Spolsky C, Uzzell T, Szymura JM. Pabijan M, et al. J Mol Evol. 2008 Sep;67(3):246-56. doi: 10.1007/s00239-008-9123-3. Epub 2008 Aug 12. J Mol Evol. 2008. PMID: 18696031 - Discovering functional DNA elements using population genomic information: a proof of concept using human mtDNA.
Schrider DR, Kern AD. Schrider DR, et al. Genome Biol Evol. 2014 Jun 9;6(7):1542-8. doi: 10.1093/gbe/evu116. Genome Biol Evol. 2014. PMID: 24916662 Free PMC article.
References
Electronic-Database Information
- Mitokor, http://www.mitokor.com/science/560mtdnasrevision.php (for the revised 560 mtDNA coding-region sequences; “zip” and “sit” files also available)
References
- Ballard JWO (2000a) Comparative genomics of mitochondrial DNA in Drosophila simulans. J Mol Evol 51:64–75 - PubMed
- ——— (2000b) Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J Mol Evol 51:48–63 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources