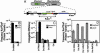Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer - PubMed (original) (raw)
Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer
Zhao-Qing Luo et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Legionella pneumophila is an intracellular pathogen that multiplies in a specialized vacuole within host cells. Biogenesis of this vacuole requires the Dot/Icm type IV protein translocation system. By using a Cre/loxP-based protein translocation assay, we found that proteins translocated by the Dot/Icm complex across the host phagosomal membrane can also be transferred from one bacterial cell to another. The flexibility of this system allowed the identification of several families of proteins translocated by the Dot/Icm complex. When analyzed by immunofluorescence microscopy, a protein identified by this procedure, SidC, was shown to translocate across the phagosomal membranes to the cytoplasmic face of the L. pneumophila phagosome. The identification of large numbers of these substrates, and the fact that the absence of any one substrate rarely results in strong defects in intracellular growth, indicate that there is significant functional redundancy among the Dot/Icm translocation targets.
Figures
Fig. 1.
Interbacterial protein translocation by the Dot/Icm system in the absence of DNA transfer. (A) Assay for interbacterial protein transfer. Translocation of Cre hybrid protein from a donor bacterial strain is measured by removal of a floxed transcriptional terminator located between the trc promoter and the npt II (kanR) gene on plasmid pZL184 harbored by a recipient bacterial strain. Bacteria harboring the intact reporter are unable to grow on media containing kanamycin and sucrose. The translocation of Cre hybrid protein into the recipient strain leads to the excision, through recombination at the loxP sites, of the DNA fragment that confers sucrose sensitivity (sacB) and reconstitution of a functional loxP-npt II translational fusion. S.D., Shine–Dalgarno sequence; npt II, neomycin phosphotransferase; red diamond, transcriptional terminator. Arrows indicate the trc promoter and loxP sites. (B) Interbacterial transfer of a fusion derived from the RSF1010 mobA gene. The mobA gene was fused to cre, and RP4-dependent protein translocation into a recipient E. coli strain was measured by using E. coli S17–1 (15) as the donor selecting kanamycin resistance and screening for gentamicin sensitivity. Black bars, S17–1 Trb+ donor; stippled bars, E. coli DH5α Trb– donor. (C). Plasmids harboring Cre fusions cannot be transferred to recipient cells. Plasmids expressing the designated proteins were harbored in either E. coli S17–1 (Trb+) or L. pneumophila Lp02 (Dot/Icm+; ref. 11), and the efficiency of plasmid transfer was measured by using either recipient E. coli or L. pneumophila strains, respectively. As a positive control, the identical plasmids having an intact oriT and mob system were used to demonstrate transfer proficiency of donor strains. Black bars, donor strain E. coli S17–1 (Trb+); gray bars, donor strain L. pneumophila Lp02 (Dot/Icm+; ref. 11). (D) Transfer of translocated Dot/Icm substrates between bacterial cells. Protein transfer was performed as described in Materials and Methods, by using either Lp02 (Dot/Icm+) or Lp03(_dotA_–) expressing the designated protein fusions as the donor strains. Gray bars, donor strain L. pneumophila Lp02 (Dot/Icm+); stippled bars, donor strain L. pneumophila Lp03 (_dotA_–).
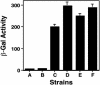
Fig. 2.
Interactions between DotF and the C termini of RalF and SidC, respectively. E. coli strain BTH101 (14) containing the indicated plasmids was grown overnight at 28°C in LB and the cultures were diluted 20-fold in the same medium containing 100 nM IPTG. Cultures were grown for 14 h before an appropriate volume was withdrawn for β-galactosidase assay. Tested strains are as follows: (column A) BTH101(pKT25dotF, pUT18C); (column B) BTH101(pKT25, pUT18CRalF(C); (column C) BTH101(pKT25dotF, pUT18CRalF(C); (column D) BTH101(pDotF (28–123), pKT25RalF(C); (column E) BTH101 [pDotF (28–123), pSidC300(C)]k; and (column F) the leucine zipper domain of the yeast GCN4 protein (14) was used as positive control in BTH101(pKT25Zip, pUT18CZip). β-galactosidase activity was expressed as Miller units. Experiments were performed in triplicate for three independent times. Data shown are from one representative experiment.
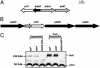
Fig. 3.
Clustering of sid genes, their paralogs into operon-like structures, and growth phase regulation of sidC.(A) sidC and its homolog sdcA are separated by only 150 bp, and the two genes are closely linked to a paralog of sidE(sdeD). (B) Three paralogs of sidE are part of a contiguous region of the chromosome that contains five significant ORFs. (C) Growth phase regulation of sidC. Bacteria were grown in AYE broth to either an OD600 = 1.8 (exponential) or 3.7 (postexponential), harvested, and analyzed by SDS/PAGE and immunoblotting with affinity-purified anti-(His)6-SidC. Displayed are Lp02(dot/icm intact; ref. 11), Lp03 (dotA_–; ref. 12), and Lp02(Δ_sdcA, Δ_sidC_), an Lp02 derivative deleted for sidC and its upstream paralog. The isocitrate dehydrogenase (ICDH) protein was used as a loading control by probing with antiserum raised against B. subtilis ICDH.
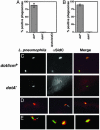
Fig. 4.
SidC is translocated by the L. pneumophila Dot/Icm system to the host cell and is localized about the phagosomal membrane. (A) Bone marrow-derived macrophages from A/J mice were infected with Lp02(dot/icm intact), Lp03(_dotA_–), or Lp02(ΔsdcA-ΔsidC) strain expressing GFP, respectively. One hour after infection, cells were fixed as described (10), and SidC was probed with anti-(His)6-SidC antibodies and Texas red-labeled secondary antibodies. Stained macrophages were scored for translocation of SidC by counting phagosomes that stained positively with anti-(His)6-SidC. Data shown are from two independent experiments performed in triplicate in which at least 100 phagosomes were scored per coverslip. (B) SidC staining on PNS prepared from _L. pneumophila_-infected U937 cells in the absence of permeabilization. Sample preparation, immunostaining, and data collection were performed as described in ref. or in A. (C) DotA-dependent translocation of SidC. (Left) Bacteria expressing GFP associated with bone marrow-derived macrophage. Strains used were Lp02(dot/icm intact; Upper) and Lp03 (_dotA_–; Lower). (Center) Immunoprobing of infected cells with anti-(His)6-SidC. (Right) Merged images of GFP and anti-(His)6-SidC staining. (D) Limited diffusion of SidC from L. pneumophila phagosome. Shown are images of Lp02(dot/icm intact) with murine bone borrow-derived macrophage. (E) SidC is translocated across the phagosomal membrane. Shown are images of PNSs of Lp02(dot/icm intact)-infected macrophages. Bacteria and SidC are probed as above, with bacteria marked by GFP and anti-SidC marked in red.
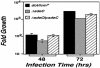
Fig. 5.
sdeC is required for efficient intracellular growth. D. discoideum cells were infected with a multiplicity of infection of 0.05, and growth of bacteria was monitored as described (13). Total bacterial cells were washed from individual microtiter wells at designated times, and the appropriate dilutions were plated on charcoal yeast extract plates to obtain the colony-forming units. Fold of growth was obtained by dividing colony-forming units at a given time point by the input bacterial cell numbers. Strains tested are as follows: black bars, Lp02(intact dot/icm); gray bars, Lp02(Δ_sdeC_); striped bars, Lp02(Δ_sdeC_) harboring pZL192 that carries the ORF of sdeC. Data shown are from two independent experiments performed in triplicate.
Similar articles
- The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation.
Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. Ninio S, et al. Mol Microbiol. 2005 Feb;55(3):912-26. doi: 10.1111/j.1365-2958.2004.04435.x. Mol Microbiol. 2005. PMID: 15661013 - A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system.
Campodonico EM, Chesnel L, Roy CR. Campodonico EM, et al. Mol Microbiol. 2005 May;56(4):918-33. doi: 10.1111/j.1365-2958.2005.04595.x. Mol Microbiol. 2005. PMID: 15853880 - The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii.
Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. Zusman T, et al. Mol Microbiol. 2007 Mar;63(5):1508-23. doi: 10.1111/j.1365-2958.2007.05604.x. Mol Microbiol. 2007. PMID: 17302824 - The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii.
Segal G, Feldman M, Zusman T. Segal G, et al. FEMS Microbiol Rev. 2005 Jan;29(1):65-81. doi: 10.1016/j.femsre.2004.07.001. FEMS Microbiol Rev. 2005. PMID: 15652976 Review. - Protein secretion in Legionella pneumophila and its relation to virulence.
Lammertyn E, Anné J. Lammertyn E, et al. FEMS Microbiol Lett. 2004 Sep 15;238(2):273-9. doi: 10.1016/j.femsle.2004.07.056. FEMS Microbiol Lett. 2004. PMID: 15358411 Review.
Cited by
- Structural characterization of the Sel1-like repeat protein LceB from Legionella pneumophila.
Penner TV, Lorente Cobo N, Patel DT, Patel DH, Savchenko A, Brassinga AKC, Prehna G. Penner TV, et al. Protein Sci. 2024 Mar;33(3):e4889. doi: 10.1002/pro.4889. Protein Sci. 2024. PMID: 38160319 Free PMC article. - Legionella hijacks the host Golgi-to-ER retrograde pathway for the association of Legionella-containing vacuole with the ER.
Kawabata M, Matsuo H, Koito T, Murata M, Kubori T, Nagai H, Tagaya M, Arasaki K. Kawabata M, et al. PLoS Pathog. 2021 Mar 24;17(3):e1009437. doi: 10.1371/journal.ppat.1009437. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33760868 Free PMC article. - Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity.
Williams SL, Schildbach JF. Williams SL, et al. Nucleic Acids Res. 2006 Jan 17;34(2):426-35. doi: 10.1093/nar/gkj444. Print 2006. Nucleic Acids Res. 2006. PMID: 16418503 Free PMC article. - Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family.
Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ. Banga S, et al. Proc Natl Acad Sci U S A. 2007 Mar 20;104(12):5121-6. doi: 10.1073/pnas.0611030104. Epub 2007 Mar 14. Proc Natl Acad Sci U S A. 2007. PMID: 17360363 Free PMC article. - Legionella pneumophila regulates host cell motility by targeting Phldb2 with a 14-3-3ζ-dependent protease effector.
Song L, Luo J, Wang H, Huang D, Tan Y, Liu Y, Wang Y, Yu K, Zhang Y, Liu X, Li D, Luo ZQ. Song L, et al. Elife. 2022 Feb 17;11:e73220. doi: 10.7554/eLife.73220. Elife. 2022. PMID: 35175192 Free PMC article.
References
- Staskawicz, B. J., Mudgett, M. B., Dangl, J. L. & Galan, J. E. (2001) Science 292, 2285–2289. - PubMed
- Kagan, J. C. & Roy, C. R. (2002) Nat. Cell Biol. 4, 945–954. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources