Yeast N-glycanase distinguishes between native and non-native glycoproteins - PubMed (original) (raw)
Yeast N-glycanase distinguishes between native and non-native glycoproteins
Christian Hirsch et al. EMBO Rep. 2004 Feb.
Abstract
N-glycanase from Saccharomyces cerevisiae (Png1) preferentially removes N-glycans from misfolded proteins. The ability of Png1 to distinguish between folded and misfolded glycoproteins is reminiscent of substrate recognition by UDP-glucose glycoprotein glucosyl transferase, an enzyme that possesses this trait. The only known in vivo substrates of Png1 are aberrant glycoproteins that originate in the endoplasmic reticulum, and arrive in the cytoplasm for proteasomal degradation. The substrate specificity of Png1 is admirably suited for this task.
Figures
Figure 1
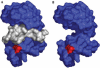
Structure of RNaseB modelled with and without S-peptide. (A) Structure of the bovine pancreatic RNaseA (Leonidas et al, 1997). The Asn34 that carries the _N_-linked glycan in the RNaseB form is rendered in red, and the S-peptide in white. (B) Model of the RNaseBS-Prot structure.
Figure 2
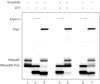
(Lanes 1–6) Digestion of native RNaseB (lane 1) with the indicated glycosidases (lanes 2–6). (Lanes 7–12) Digestion of reduced and alkylated RNaseB (lane 7) with the indicated glycosidases. After enzymatic digestion, samples were analysed by SDS–PAGE followed by silver staining. The number of glycans is indicated on the right. The following amounts of enzyme were used: Endo A: 1,440 U; Endo H: 100 U; Png F: 5,000 U; Png1: 420 U. The total amount of substrate (RNaseB) for each reaction volume was 0.4 μg in 50 μl. Samples were incubated at 20–23°C for 16 h.
Figure 3
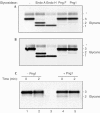
RNaseB was partially digested with subtilisin, yielding a preparation that contained undigested RNaseB and the faster migrating RNaseBS-Prot (lane 1). Digestion with Endo H deglycosylated both forms of the RNaseB (lane 2), whereas Png1 deglycosylated only the RNaseBS-Prot (lane 3). RNaseB reconstituted with S-peptide is susceptible to deglycosylation by Endo H (lane 4), but resistant to deglycosylation by Png1 (lane 5). The activity of Endo H is not reduced in the absence of DTT (lane 6), whereas Png1 activity shows a marked reduction in activity (lane 7). Endo H: 25 U; Png1: 42 U. Total RNaseB concentration was 5 μg in a final volume of 30 μl. Reactions were incubated at 20–23°C for 16 h.
Figure 4
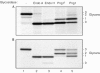
Deglycosylation profile of α1-AT from HepG2 and HEK-293 cells. (A) α1-AT was recovered by immunoprecipitation from 35S-methionine-labelled HepG2 extracts that were either left untreated (lane 1) or incubated with the indicated glycosidases (lanes 2–5). Immunoprecipitates were analysed by SDS–PAGE followed by fluorography. The asterisk (*) indicates _N_-linked glycans of the complex type. (B) As in (A), the source of the α1-AT was transiently transfected HEK-293 cells. (C) HEK-293 cells transiently transfected with wt-α1-AT were pulse labelled with 35S-methionine for 1 min and then lysed either 0 or 2 min after the chase. Samples were left untreated (lanes 1 and 2) or incubated with Png1 (lanes 4 and 5). Endo A: 2,400 U; Endo H: 1,500 U; Png F: 5,000 U; Png1: 2,800 U. Reactions were preformed in a total volume of 1 ml, incubated 16 h at 4°C.
Figure 5
Mutants of α1-AT are more susceptible to deglycosylation by Png1. (A) HEK-293 cells were transiently transfected with NHK-AT. NHK-AT was recovered by immunoprecipitation from 35S-methionine-labelled lysates that were either left untreated (lane 1) or incubated with the indicated glycosidases (lanes 2–5). Immunoprecipitates were analysed by SDS–PAGE followed by fluorography. (B) Similar set of experiments as in (A), the substrate used here is Pi Z-AT. Endoglycosidase concentrations were the same as in Fig 4.
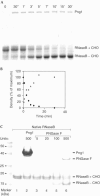
Figure 6
Kinetics of Png1. (A) Rate of deglycosylation of RNaseB by Png1. Png1 (0.56 nM) was incubated with RNaseB (0.122 mM) at 30°C. Samples were taken at the indicated time points and subjected to SDS–PAGE followed by Coomassie staining. The zero time point was taken prior to addition of Png1. (B) Quantitation of RNaseB deglycosylation by densitometry. The glycosylated form of RNaseB (+CHO; substrate, black diamonds) and the deglycosylated form of RNaseB (−CHO; product, grey squares) are indicated. (C) Png1 does not attack native RNaseB, while PNGase F recognizes and deglycosylates native RNaseB. Native RNaseB was incubated with the indicated units of Png1 or PNGase F at 20–23°C for 16 h, and fractions were subjected to SDS–PAGE, followed by Coomassie staining. Even at high enzyme concentrations, Png1 fails to deglycosylate native RNaseB.
Similar articles
- Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae.
Hirayama H, Seino J, Kitajima T, Jigami Y, Suzuki T. Hirayama H, et al. J Biol Chem. 2010 Apr 16;285(16):12390-404. doi: 10.1074/jbc.M109.082081. Epub 2010 Feb 11. J Biol Chem. 2010. PMID: 20150426 Free PMC article. - Influence of substrate conformation on the deglycosylation of ribonuclease B by recombinant yeast peptide:N-glycanase.
Wang S, Wang PG, Qi Q. Wang S, et al. Acta Biochim Biophys Sin (Shanghai). 2007 Jan;39(1):8-14. doi: 10.1111/j.1745-7270.2007.00244.x. Acta Biochim Biophys Sin (Shanghai). 2007. PMID: 17213953 - N-terminal deletion of peptide:N-glycanase results in enhanced deglycosylation activity.
Wang S, Xin F, Liu X, Wang Y, An Z, Qi Q, Wang PG. Wang S, et al. PLoS One. 2009 Dec 16;4(12):e8335. doi: 10.1371/journal.pone.0008335. PLoS One. 2009. PMID: 20016784 Free PMC article. - Physiological and molecular functions of the cytosolic peptide:N-glycanase.
Hirayama H, Hosomi A, Suzuki T. Hirayama H, et al. Semin Cell Dev Biol. 2015 May;41:110-20. doi: 10.1016/j.semcdb.2014.11.009. Epub 2014 Dec 2. Semin Cell Dev Biol. 2015. PMID: 25475175 Review. - The cytoplasmic peptide:N-glycanase (Ngly1)-basic science encounters a human genetic disorder.
Suzuki T. Suzuki T. J Biochem. 2015 Jan;157(1):23-34. doi: 10.1093/jb/mvu068. Epub 2014 Nov 13. J Biochem. 2015. PMID: 25398991 Review.
Cited by
- Development of new NGLY1 assay systems - toward developing an early screening method for NGLY1 deficiency.
Hirayama H, Fujihira H, Suzuki T. Hirayama H, et al. Glycobiology. 2024 Sep 30;34(11):cwae067. doi: 10.1093/glycob/cwae067. Glycobiology. 2024. PMID: 39206713 Free PMC article. Review. - A congenital disorder of deglycosylation: Biochemical characterization of N-glycanase 1 deficiency in patient fibroblasts.
He P, Grotzke JE, Ng BG, Gunel M, Jafar-Nejad H, Cresswell P, Enns GM, Freeze HH. He P, et al. Glycobiology. 2015 Aug;25(8):836-44. doi: 10.1093/glycob/cwv024. Epub 2015 Apr 21. Glycobiology. 2015. PMID: 25900930 Free PMC article. - Deglycosylated milin unfolds via inactive monomeric intermediates.
Yadav SC, Prasanna Kumari NK, Jagannadham MV. Yadav SC, et al. Eur Biophys J. 2010 Nov;39(12):1581-8. doi: 10.1007/s00249-010-0615-x. Epub 2010 Jun 13. Eur Biophys J. 2010. PMID: 20549500 - Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator.
Chang XB, Mengos A, Hou YX, Cui L, Jensen TJ, Aleksandrov A, Riordan JR, Gentzsch M. Chang XB, et al. J Cell Sci. 2008 Sep 1;121(Pt 17):2814-23. doi: 10.1242/jcs.028951. Epub 2008 Aug 5. J Cell Sci. 2008. PMID: 18682497 Free PMC article. - HIV-1 envelope resistance to proteasomal cleavage: implications for vaccine induced immune responses.
Steers NJ, Ratto-Kim S, de Souza MS, Currier JR, Kim JH, Michael NL, Alving CR, Rao M. Steers NJ, et al. PLoS One. 2012;7(8):e42579. doi: 10.1371/journal.pone.0042579. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880042 Free PMC article.
References
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987) Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506–512 - PubMed
- Fabbretti G, Sergi C, Consales G, Faa G, Brisigotti M, Romeo G, Callea F (1992) Genetic variants of alpha-1-antitrypsin (AAT). Liver 12: 296–301 - PubMed
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K (2003) Enhancement of endoplasmic reticulum (ER) degradation of misfolded null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem 278: 26287–26294 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases