Extensive maternal DNA hypomethylation in the endosperm of Zea mays - PubMed (original) (raw)
Extensive maternal DNA hypomethylation in the endosperm of Zea mays
Massimiliano Lauria et al. Plant Cell. 2004 Feb.
Abstract
A PCR-based genomic scan has been undertaken to estimate the extent and ratio of maternally versus paternally methylated DNA regions in endosperm, embryo, and leaf of Zea mays (maize). Analysis of several inbred lines and their reciprocal crosses identified a large number of conserved, differentially methylated DNA regions (DMRs) that were specific to the endosperm. DMRs were hypomethylated at specific methylation-sensitive restriction sites upon maternal transmission, whereas upon paternal transmission, the methylation levels were similar to those observed in embryo and leaf. Maternal hypomethylation was extensive and offers a likely explanation for the 13% reduction in methyl-cytosine content of the endosperm compared with leaf tissue. DMRs showed identity to expressed genic regions, were observed early after fertilization, and maintained at a later stage of endosperm development. The implications of extensive maternal hypomethylation with respect to endosperm development and epigenetic reprogramming will be discussed.
Figures
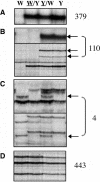
Figure 1.
Identification of DMRs in Endosperm DNA by MSAP. Representative MSAP profiles of an EcoRI/HpaII digest of endosperm DNA extracted from the inbred lines W64A (W) and A69Y (Y) and the reciprocal crosses
W64
/A69Y (
W
/Y) and
A69Y
/W64 (
Y
/W) (the seed parent of the reciprocal cross is underlined) harvested at 15 DAP. The sum of profiles scored of both inbred lines using 24 selective primer combinations is indicated at the right of each panel. (A) An MSAP restriction fragment specific to the A69Y inbred line that did not show parental differences in methylation (a normal polymorphic profile). (B) MSAP restriction fragments that only were detected when the A69Y inbred line was the seed parent (maternal profiles are indicated by arrows). (C) MSAP restriction fragments that only were detected when the W64A inbred line was the pollen parent (paternal profiles are indicated by arrows). (D) Monomorphic profiles.
Figure 2.
Analysis of DMRs in Different Z. mays Inbred Lines by MSAP. Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from the inbred lines W64A, A69Y, and W23 (23) and their respective reciprocal crosses harvested at 15 DAP. Other abbreviations are as in Figure 1. (A) MMPs specific to the W23 and A69Y inbred lines (top panel) or to the W64A inbred line (bottom panel) are indicated by arrows. (B) An MMP that was conserved between the W64 and W23 inbred lines (top panel) and an MMP that was conserved between the A69Y and W23 inbred lines (bottom panel).
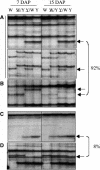
Figure 3.
Stability of DMRs during Endosperm Development by MSAP. Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from endosperms harvested at 7 and 15 DAP from the W64A and A69Y inbred lines and their reciprocal crosses. The percentage of DMRs with a given profile is indicated at the right. This percentage was calculated from profiles scored of two inbred lines with 16 selective primer combinations. Arrows indicate DMRs, and abbreviations are as in Figure 1. (A) An MMP and a PMP that were stable at the two developmental stages studied (top and bottom panels, respectively). (B) An MMP that was stable at the two developmental stages, albeit with a lower intensity at 15 DAP. (C) A restriction fragment that showed an MMP at 7 DAP but a normal polymorphic profile at 15 DAP. (D) A restriction fragment that showed an MMP at 7 DAP but a monomorphic profile at 15 DAP.
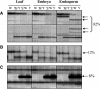
Figure 4.
Analysis of DMRs in Leaf, Embryo, and Endosperm Tissues by MSAP. Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from 2-week-old leaf tissues and from embryos and endosperm harvested at 15 DAP of the inbred lines W64A, A69Y, and their reciprocal crosses. The percentage of MMPs that exhibited each profile is indicated at the right. Arrows indicate MMPs, and abbreviations are as in Figure 1. (A) Restriction fragments that showed an MMP specific to the endosperm. (B) A restriction fragment that was detected in all tissues but only showed an MMP in the endosperm. (C) A restriction fragment that showed an MMP in all tissues (a common MMP).
Figure 5.
Reciprocal Backcross Analysis of DMRs in Endosperm Tissue by MSAP. Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from 18 individual endosperms of the reciprocal backcrosses (
W64A/A69Y
)/W64A and (
A69Y/W64A
)/W64A, indicated as (
W/Y
)/W and (
Y/W
)/W, respectively) harvested at 15 DAP. The MSAP profiles of the inbred lines W64A and A69Y and their respective reciprocal F1 hybrids are shown in the panels at right. Arrows indicate DMRs specific to the A69Y inbred line. Other abbreviations are as in Figure 1. (A) A restriction fragment representing a common MMP that showed a grandparental effect in the reciprocal backcrosses. (B) A restriction fragment representing an endosperm-specific MMP that segregated in both backcrosses. (C) A restriction fragment representing an endosperm-specific PMP that was not detected upon maternal transmission.
Figure 6.
Parental Methylation Status of a Restriction Fragment That Showed an MMP by MSAP. An EcoRI (E)/HpaII (H) restriction fragment showing the possible methylation status of HpaII sites upon maternal versus paternal transmission. The asterisks indicate a methylated cytosine residue. (A) A fragment without internal HpaII sites. The external HpaII site is maternally hypomethylated and paternally methylated. (B) A fragment with an internal HpaII site. The external and internal HpaII sites are maternally hypomethylated and hypermethylated, respectively. The methylation status of this paternal fragment may be one of the following: i, hypomethylated at both HpaII sites; ii, hypomethylated at the internal HpaII site only; or iii, hypermethylated at both HpaII sites.
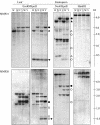
Figure 7.
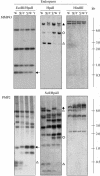
DNA Gel Blot Analysis of DMRs. DNA extracted from endosperms harvested at 15 DAP or from 2-week-old leaves of the inbred lines W64A, A69Y, and their reciprocal hybrids was digested with the restriction enzymes indicated. DNA gel blot analysis of leaf and endosperm DNA probed with MMP21 and MMP26 that lacked internal HpaII sites (top and bottom panels, respectively). MMP21 was a 309-bp fragment that showed an MMP specific to the W64A inbred line by MSAP; MMP26 was a 509-bp fragment that exhibited an MMP specific to the A69Y inbred line by MSAP. Arrows indicate a fragment that corresponds in size to the DMR isolated by MSAP. Open and closed triangles indicate MMPs and PMPs, respectively, specific to the W64A inbred line, and open and closed circles indicate MMPs and PMPs, respectively, specific to the A69Y inbred line. Abbreviations are as in Figure 1.
Figure 8.
DNA Gel Blot Analysis of DMRs. DNA extracted from endosperms harvested at 15 DAP or from 2-week-old leaves of the inbred lines W64A, A69Y, and their reciprocal hybrids was digested with the restriction enzymes indicated. DNA gel blot analysis of endosperm DNA probed with MMP43 and PMP2 that contained an internal HpaII site (top and bottom panels, respectively). MMP43 was a 665-bp fragment that showed an MMP specific to the W64A inbred line by MSAP; PMP2 was a 985-bp fragment that exhibited a PMP specific to the W64A inbred line by MSAP. Open and closed triangles indicate MMPs and PMPs, respectively, specific to the W64A inbred line, and open and closed circles indicate MMPs and PMPs, respectively, specific to the A69Y inbred line. Arrows indicate a fragment that corresponds in size to the DMR isolated by MSAP. Abbreviations are as in Figure 1.
Similar articles
- Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may).
Wang P, Xia H, Zhang Y, Zhao S, Zhao C, Hou L, Li C, Li A, Ma C, Wang X. Wang P, et al. BMC Genomics. 2015 Jan 23;16(1):21. doi: 10.1186/s12864-014-1204-7. BMC Genomics. 2015. PMID: 25612809 Free PMC article. - Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L.
Lund G, Ciceri P, Viotti A. Lund G, et al. Plant J. 1995 Oct;8(4):571-81. doi: 10.1046/j.1365-313x.1995.8040571.x. Plant J. 1995. PMID: 7496402 - Regulation of aleurone development in cereal grains.
Becraft PW, Yi G. Becraft PW, et al. J Exp Bot. 2011 Mar;62(5):1669-75. doi: 10.1093/jxb/erq372. Epub 2010 Nov 25. J Exp Bot. 2011. PMID: 21109580 Review. - Genomic imprinting: insights from plants.
Gehring M. Gehring M. Annu Rev Genet. 2013;47:187-208. doi: 10.1146/annurev-genet-110711-155527. Epub 2013 Aug 30. Annu Rev Genet. 2013. PMID: 24016190 Review.
Cited by
- The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization.
Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U. Baroux C, et al. Plant Cell. 2007 Jun;19(6):1782-94. doi: 10.1105/tpc.106.046235. Epub 2007 Jun 8. Plant Cell. 2007. PMID: 17557811 Free PMC article. - Reprogramming the epigenome during germline and seed development.
Johnson MA, Bender J. Johnson MA, et al. Genome Biol. 2009;10(8):232. doi: 10.1186/gb-2009-10-8-232. Epub 2009 Aug 24. Genome Biol. 2009. PMID: 19725931 Free PMC article. Review. - Temporal changes in transcripts of miniature inverted-repeat transposable elements during rice endosperm development.
Nagata H, Ono A, Tonosaki K, Kawakatsu T, Sato Y, Yano K, Kishima Y, Kinoshita T. Nagata H, et al. Plant J. 2022 Mar;109(5):1035-1047. doi: 10.1111/tpj.15698. Plant J. 2022. PMID: 35128739 Free PMC article. - Control of transposable elements in Arabidopsis thaliana.
Ito H, Kakutani T. Ito H, et al. Chromosome Res. 2014 Jun;22(2):217-23. doi: 10.1007/s10577-014-9417-9. Chromosome Res. 2014. PMID: 24801341 Review. - Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may).
Wang P, Xia H, Zhang Y, Zhao S, Zhao C, Hou L, Li C, Li A, Ma C, Wang X. Wang P, et al. BMC Genomics. 2015 Jan 23;16(1):21. doi: 10.1186/s12864-014-1204-7. BMC Genomics. 2015. PMID: 25612809 Free PMC article.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127, 2493–2502. - PubMed
- Alleman, M., and Doctor, J. (2000). Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol. Biol. 43, 147–161. - PubMed
- Baroux, C., Spillane, C., and Grossniklaus, U. (2002). Genomic imprinting during seed development. Adv. Genet. 46, 165–214. - PubMed
- Bianchi, M.W., and Viotti, A. (1988). DNA methylation and tissue-specific transcription of the storage protein gene of maize. Plant Mol. Biol. 11, 203–214. - PubMed
- Birchler, J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27, 181–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources