AIRE functions as an E3 ubiquitin ligase - PubMed (original) (raw)
. 2004 Jan 19;199(2):167-72.
doi: 10.1084/jem.20031291.
Shigetsugu Hatakeyama, Akemi Matsushima, Hongwei Han, Satoshi Ishido, Hak Hotta, Jun Kudoh, Nobuyoshi Shimizu, Vassilis Doucas, Keiichi I Nakayama, Noriyuki Kuroda, Mitsuru Matsumoto
Affiliations
- PMID: 14734522
- PMCID: PMC2211764
- DOI: 10.1084/jem.20031291
AIRE functions as an E3 ubiquitin ligase
Daisuke Uchida et al. J Exp Med. 2004.
Abstract
Autoimmune regulator (AIRE) gene mutation is responsible for the development of autoimmune-polyendocrinopathy-candidiasis ectodermal dystrophy, an organ-specific autoimmune disease with monogenic autosomal recessive inheritance. AIRE is predominantly expressed in medullary epithelial cells of the thymus and is considered to play important roles in the establishment of self-tolerance. AIRE contains two plant homeodomain (PHD) domains, and the novel role of PHD as an E3 ubiquitin (Ub) ligase has just emerged. Here we show that the first PHD (PHD1) of AIRE mediates E3 ligase activity. The significance of this finding was underscored by the fact that disease-causing missense mutations in the PHD1 (C311Y and P326Q) abolished its E3 ligase activity. These results add a novel enzymatic function for AIRE and suggest an indispensable role of the Ub proteasome pathway in the establishment of self-tolerance, in which AIRE is involved.
Figures
Figure 1.
AIRE mediates E3 ligase activity through PHD1. (A) The PHD1 and PHD2 domains of AIRE showed homology with the PHDs from other proteins showing E3 ligase activity. *, the position of disease-causing mutations (C311Y and P326Q) used in the study. (B) The GST-PHD1, but not GST-PHD2, fusion protein exhibited polyubiquitylated proteins when rabbit reticulocyte lysate was used for in vitro ubiquitylation assays (top). SDS-PAGE of GST-fusion proteins used in the assay (10% input) is shown on the bottom. (C) Recombinant E1 and E2 (Ubc4) were used for in vitro ubiquitylation assays (top). The ubiquitylation reactions were subjected to immunoblot analysis with antibodies against GST (bottom). E3 ligase activity mediated through PHD1 is incubation dependent. *, nonspecific bands. (D) The full-sized recombinant AIRE protein was successfully and homogeneously expressed in Sf21 cells using a baculovirus expression system. The purified recombinant AIRE protein was subjected to immunoblot analysis with an antibody against His6-tag. (E) Ubc4 E2 preference by the full-sized recombinant AIRE in in vitro ubiquitylation assays (top). CHIP, a U-box–type E3 ligase, is included to verify the specificity and biological activities of E2s (middle; reference 22). Equal amount of E2s were used in the reaction (bottom). (F) Specificity of E3 ligase activity mediated by the full-sized recombinant AIRE in in vitro ubiquitylation assays.
Figure 2.
Mutations in PHD1 affect E3 ligase activity. (A) Schematic representation of the PHD mutants used in the study. (B) Substitutions of the first cysteines into alanines in PHD1, or PHD1 and PHD2, but not in PHD2 alone, abolished E3 ligase activity (top). The ubiquitylation reactions were subjected to immunoblot analysis with antibodies against His6-tag (bottom). (C) Disease-causing mutations of PHD1 (C311Y and P326Q) resulted in loss of E3 ligase activity as assessed with in vitro ubiquitylation assays.
Figure 3.
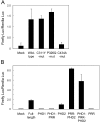
Effect of PHD mutations on the transcriptional transactivating properties of AIRE. (A) Various forms of full-length AIRE including disease-causing mutations were fused with Gal4-binding domain and transfected into mTEC cells together with a reporter plasmid. Firefly luciferase activities were normalized with respect to the Renilla luciferase activities used for assessment of the transfection efficiency. No significant reduction of Gal4 activation was observed from the disease-causing mutations of PHD1 (C311Y and P326Q), whereas the PHD2 mutant (C434A) showed loss of transcriptional activation. The results are expressed as the mean ± SEM for triplicate wells during one representative experiment from a total of five repeat experiments. (B) Isolated PHDs were tested for the transcriptional activation with the same system described in A. The fragments used are as follows (see Figs. 1 A and 2 A): PHD1, amino acids 292–341; PHD1 plus PRR, 292–432; PHD2, 433–545; PRR plus PHD2, 342–545; PHD1 plus PRR plus PHD2, 292–545; and PRR, 342–432. PHD2 suffices for the transcriptional activation in this assay, whereas PHD1 has no such activities. The results are expressed as the mean ± SEM for triplicate wells during one representative experiment from a total of three repeat experiments.
Similar articles
- APECED-causing mutations in AIRE reveal the functional domains of the protein.
Halonen M, Kangas H, Rüppell T, Ilmarinen T, Ollila J, Kolmer M, Vihinen M, Palvimo J, Saarela J, Ulmanen I, Eskelin P. Halonen M, et al. Hum Mutat. 2004 Mar;23(3):245-57. doi: 10.1002/humu.20003. Hum Mutat. 2004. PMID: 14974083 - NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease.
Bottomley MJ, Stier G, Pennacchini D, Legube G, Simon B, Akhtar A, Sattler M, Musco G. Bottomley MJ, et al. J Biol Chem. 2005 Mar 25;280(12):11505-12. doi: 10.1074/jbc.M413959200. Epub 2005 Jan 13. J Biol Chem. 2005. PMID: 15649886 - Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutation present with only a mild autoimmune phenotype.
Hubert FX, Kinkel SA, Crewther PE, Cannon PZ, Webster KE, Link M, Uibo R, O'Bryan MK, Meager A, Forehan SP, Smyth GK, Mittaz L, Antonarakis SE, Peterson P, Heath WR, Scott HS. Hubert FX, et al. J Immunol. 2009 Mar 15;182(6):3902-18. doi: 10.4049/jimmunol.0802124. J Immunol. 2009. PMID: 19265170 - AIRE and immunological tolerance: insights from the study of autoimmune polyendocrinopathy candidiasis and ectodermal dystrophy.
Notarangelo LD, Mazza C, Forino C, Mazzolari E, Buzi F. Notarangelo LD, et al. Curr Opin Allergy Clin Immunol. 2004 Dec;4(6):491-6. doi: 10.1097/00130832-200412000-00004. Curr Opin Allergy Clin Immunol. 2004. PMID: 15640689 Review. - AIRE and APECED: molecular insights into an autoimmune disease.
Villaseñor J, Benoist C, Mathis D. Villaseñor J, et al. Immunol Rev. 2005 Apr;204:156-64. doi: 10.1111/j.0105-2896.2005.00246.x. Immunol Rev. 2005. PMID: 15790357 Review.
Cited by
- Chromosomal clustering of genes controlled by the aire transcription factor.
Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP, Benoist CO, Mathis DJ. Johnnidis JB, et al. Proc Natl Acad Sci U S A. 2005 May 17;102(20):7233-8. doi: 10.1073/pnas.0502670102. Epub 2005 May 9. Proc Natl Acad Sci U S A. 2005. PMID: 15883360 Free PMC article. - Structures of protein domains that create or recognize histone modifications.
Bottomley MJ. Bottomley MJ. EMBO Rep. 2004 May;5(5):464-9. doi: 10.1038/sj.embor.7400146. EMBO Rep. 2004. PMID: 15184976 Free PMC article. Review. - Does antigen masking by ubiquitin chains protect from the development of autoimmune diseases?
Weil R. Weil R. Front Immunol. 2014 Jun 3;5:262. doi: 10.3389/fimmu.2014.00262. eCollection 2014. Front Immunol. 2014. PMID: 24917867 Free PMC article. Review. - TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism.
Brigant B, Metzinger-Le Meuth V, Rochette J, Metzinger L. Brigant B, et al. Int J Mol Sci. 2018 Dec 24;20(1):67. doi: 10.3390/ijms20010067. Int J Mol Sci. 2018. PMID: 30586926 Free PMC article. Review. - Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription.
Žumer K, Plemenitaš A, Saksela K, Peterlin BM. Žumer K, et al. Nucleic Acids Res. 2011 Oct;39(18):7908-19. doi: 10.1093/nar/gkr527. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724609 Free PMC article.
References
- Kamradt, T., and N.A. Mitchison. 2001. Tolerance and autoimmunity. N. Engl. J. Med. 344:655–664. - PubMed
- Wanstrat, A., and E. Wakeland. 2001. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat. Immunol. 2:802–809. - PubMed
- Nagamine, K., P. Peterson, H.S. Scott, J. Kudoh, S. Minoshima, M. Heino, K.J. Krohn, M.D. Lalioti, P.E. Mullis, S.E. Antonarakis, et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17:393–398. - PubMed
- The Finnish-German APECED Consortium. 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17:399–403. - PubMed
- Björses, P., J. Aaltonen, N. Horelli-Kuitunen, M.L. Yaspo, and L. Peltonen. 1998. Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 7:1547–1553. - PubMed