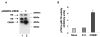Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene - PubMed (original) (raw)
Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene
Sarmila Majumder et al. J Biol Chem. 2004.
Retraction in
- Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene.
Majumder S, Varadharaj S, Ghoshal K, Monani U, Burghes AHM, Jacob ST. Majumder S, et al. J Biol Chem. 2018 Aug 17;293(33):12946. doi: 10.1074/jbc.W118.004968. J Biol Chem. 2018. PMID: 30120150 Free PMC article. No abstract available.
Abstract
Spinal muscular atrophy, an autosomal recessive disorder, is caused by loss of the SMN1 (survival motor neuron) gene while retaining the SMN2 gene. SMN1 produces a majority of full-length SMN transcript, whereas SMN2 generates mostly an isoform lacking exon 7. Here, we demonstrate a novel cAMP-response element, CRE-II, in the SMN promoter that interacts with the cAMP-response element-binding (CREB) family of proteins. In vitro DNase I protection analysis and in vivo genomic footprinting of the SMN promoter using the brain and liver nuclei from SMN2 transgenic mice revealed footprinting at the CRE-II site. Site-directed mutation of the CRE-II element caused a marked reduction in the SMN promoter activity revealed by transient transfection assay. Activation of the cAMP pathway by dibutyryl cAMP (0.5 mm) alone or in combination with forskolin (20 microm) caused a 2-5-fold increase in the SMN promoter activity but had no effect on the CRE-II mutated promoter. Electrophoretic mobility shift assay and a UV-induced DNA-protein cross-linking experiment confirmed that CREB1 binds specifically to the CRE-II site. Transient overexpression of CREB1 protein resulted in a 4-fold increase of the SMN promoter activity. Intraperitoneal injection of epinephrine in mice expressing two copies of the human SMN2 gene resulted in a 2-fold increase in full-length SMN transcript in the liver. Combined treatment with dibutyryl cAMP and forskolin significantly increased the level of both the full-length and exon 7-deleted SMN (exonDelta7SMN) transcript in primary hepatocytes from mice expressing two copies of human SMN2 gene. Similar treatments of type I spinal muscular atrophy mouse and human fibroblasts as well as HeLa cells resulted in an augmented level of SMN transcript. These findings suggest that the CRE-II site in SMN promoter positively regulates the expression of the SMN gene, and treatment with cAMP-elevating agents increases expression of both the full-length and exonDelta7SMN transcript.
Figures
Fig. 1
A, schematic diagram of 750-bp SMN2 promoter region depicting relevant cis-elements. The arrows indicate the positions of selected restriction sites. The SstI/StyI, StyI/PstI, and PstI/HindIII probes represent the fragments of the SMN2 promoter that were subsequently used for DNase I footprinting studies. B, sequence of the CRE consensus, CRE-I and -II elements, and the mutant CRE-II of the SMN2 promoter.
Fig. 2. In vivo genomic footprinting demonstrates involvement of CRE-II site in SMN2 gene expression
Intact nuclei isolated from brain and liver cells of _Smn_−/− mice expressing eight copies of human SMN2 gene were exposed to limited dimethylsulfate treatment, and genomic DNA was isolated. The DNA was then subjected to piperidine treatment followed by LM-PCR amplification of the SMN2 promoter. The LM-PCR products were separated on 6% sequencing gel and exposed to x-ray film. N, naked DNA, where DNA was treated with dimethyl sulfate and piperidine after isolation; C, DNA isolated from control cells treated in vivo with dimethyl sulfate. The stars and arrows indicate hypersensitive and protected G residues, respectively. A, lower strand spanning from +210 to +283 bp. B, upper strand spanning from −312 to −443 bp of the SMN2 promoter.
Fig. 3. DNase I footprinting reveals protection of CRE-II site in SMN proximal promoter
A, a radiolabeled probe was generated by PstI/HindIII digestion of human SMN gene promoter spanning the region between +150 to +300 bp. The upper strand of the DNA fragment was labeled with [γ-32P]ATP and T4 polynucleotide kinase. HeLa nuclear extract was allowed to interact with the labeled probe and subjected to limited DNase I digestion. For competition assay, an unlabeled PstI/HindIII fragment was added to the reaction prior to the addition of labeled probe. Lane 1, free probe; lane 2, DNA ladder, no extract; lanes 3–5, 25, 50, and 75 μg of HeLa nuclear extract, respectively; lanes 6 and 7, 75 μg of HeLa nuclear extract in presence of a 50- and 100-fold excess of unlabeled PstI/HindIII fragment, respectively. B, probe corresponding to the noncoding strand of the PstI/HindIII fragment was labeled using Klenow and [α-32P]dGTP. Lane 1, free probe; lane 2, DNA ladder, no extract; lanes 3–6, 25, 50, 75, and 100 μg of HeLa nuclear extract; lanes 7 and 8, 100 μg of HeLa nuclear extract in the presence of a 50- and 100-fold excess of unlabeled PstI/HindIII fragment, respectively. A/G lane, A + G ladder of the probe.
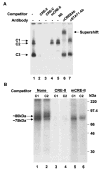
Fig. 4. EMSA and UV-cross-linking study shows binding of CREB-1 protein to the CRE-II site of the SMN proximal promoter
A, DNA mobility shift and supershift assays were performed using [γ-32P]ATP end-labeled CRE-II oligonucleotide. One nanogram of 32P-CRE-II and 10 μg of EHMN nuclear extract (NE) was used in each reaction. Lane 1, nuclear extract only; lane 2, 200× CRE-II oligonucleotide; lane 3, 200× CRE consensus oligonucleotide (CRE-C); lane 4, 200× mutant CRE-II oligonucleotide (mCRE-II); lane 5, 200× nonspecific oligonucleotide (N.S.); lane 6, anti-CREB/ATF-1 antibody (monoclonal); lane 7, anti-STAT1 antibody. B, EHMN nuclear extract and 32P-labeled CRE-II oligonucleotide were allowed to form a complex in EMSA binding buffer, separated on 5% acrylamide gel, and the protein-DNA was cross-linked under UV light. The C1 and C2 complexes were recovered from the gel, eluted, and run on 10% SDS-PAGE. Lanes 1, 3, and 5, complex C2 recovered from control reaction and reaction mixtures containing excess CRE-II and mCRE-II oligonucleotide, respectively. Lanes 2, 4, and 6, complex C1 recovered from control reaction and reaction mixtures containing excess CRE-II and mCRE-II oligonucleotide, respectively.
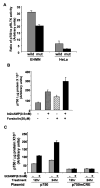
Fig. 5
A, transient transfection assay demonstrates importance of the CRE-II site in SMN promoter activity. EHMN and HeLa cells were plated at 1 × 105 cells/well in 6-well dishes and transfected with either 150 or 500 ng, respectively, of p750 (wild type; wild) or p750mCRE (mutant; mut) and 40 ng of pRLTK (internal control) plasmid DNA. Cell extracts were prepared in 1× lysis buffer (Promega), 48 h post-transfection, and luciferase activity was measured using the dual luciferase assay kit. The promoter activity is presented as the ratio of p750/pRLTK activity. B, both Bt2cAMP and forskolin up-regulates SMN promoter activity. HeLa cells were transiently transfected with 4 μg of p750 plasmid and were treated with different concentrations ofBt2cAMP and/or forskolin 24 h post-transfection as indicated. Cells were harvested 24 h after the respective treatment and measured for luciferase activity. The promoter activity is presented as the p750 activity/μg of protein. C, mutation at the CRE-II sequence abolishes cAMP responsiveness of the promoter. HeLa cells were transfected with plasmid p750 or p750mCRE. The cells were treated with 0.5 mM Bt2cAMP 24 h post-transfection and harvested after 12 and 24 h of treatment. Luciferase activity was measured as described, and values are expressed as per μg of protein. All of the data are representative of three independent experiments ± S.E.
Fig. 6. Transient overexpression of CREB-1 protein up-regulates SMN promoter activity
A, HeLa cells were transiently transfected with empty vector (lane 1) or CREB-1 overexpression vector (pSGRSV-CREB) (lane 2). Whole cell extract prepared 48 h post-transfection was subjected to Western blot analysis with anti-CREB/ATF-1 antibody. B, HeLa cells were co-transfected with plasmids p750, internal control pRLTK, and either pSGRSV-CREB (CREB) or the corresponding empty vector (E.V.). Cells were harvested 48 h post-transfection, and luciferase activity was measured using the dual luciferase assay kit. The promoter activity was expressed as a ratio of p750 to pRLTK activity. The data are representative of three independent experiments ± S.E.
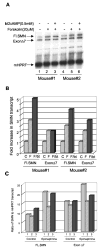
Fig. 7. cAMP-elevating agents stimulate expression of SMN transcripts in mouse primary hepatocytes as well as in mouse liver
A, primary hepatocytes isolated from Smn+/− mice expressing two copies of the human SMN2 gene were treated with Bt2cAMP and/or forskolin for 8 h. Total RNA isolated from untreated and treated cells were subjected to multiplex PCR, and the products are separated on a sequencing gel. The experiment was done with hepatocytes isolated from two different mice. FL.SMN, full-length SMN. B, for quantitation of the mouse HPRT transcript and different splice variants of SMN transcripts, the dried gel was exposed to storage phosphor screen (Amersham Biosciences) for different length of time and analyzed using ImageQuant software. The ratio of SMN transcript to HPRT transcript was calculated, and data are expressed as -fold increase in SMN transcript compared with the untreated control taken as 1. The increase in SMN full-length transcript is 4.5 ± 0.7-fold in presence of forskolin and Bt2cAMP compared with the untreated control. C, human SMN2 transgenic mice (in triplicate) were injected intraperitoneally with epinephrine every 2 h for 6 h and sacrificed 2 h after the last injection. Total RNA isolated from the liver was analyzed for SMN gene transcription by multiplex PCR as mentioned above. After quantitation of the PCR product, the data are presented as ratio of SMN transcript to that of HPRT for an individual mouse (–3). The level of SMN full-length transcript in untreated mice is 9.6 ± 2.0, and the level in epinephrine treated mice is 18 ± 4.3.
Fig. 8. Forskolin and Bt2cAMP up-regulates expression of both the full-length and exon 7-deleted SMN transcript
A, mouse type I SMA fibroblasts were treated with Bt2cAMP and/or forskolin for 8 h. Total RNA isolated from untreated and treated cells were subjected to multiplex PCR, and the products are separated on a sequencing gel. B, alteration in the transcript level of different SMN isoforms were quantitated using ImageQuant software and represented as a ratio of SMN to HPRT (internal control) transcript level. C, HeLa cells were subjected to similar treatment as described for mouse fibroblasts and analyzed by multiplex PCR. D, bar diagram representing the effect of Bt2cAMP and/or forskolin on different SMN isoforms in HeLa cells.
Similar articles
- Therapeutics development for spinal muscular atrophy.
Sumner CJ. Sumner CJ. NeuroRx. 2006 Apr;3(2):235-45. doi: 10.1016/j.nurx.2006.01.010. NeuroRx. 2006. PMID: 16554261 Free PMC article. Review. - Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy.
Thurmond J, Butchbach ME, Palomo M, Pease B, Rao M, Bedell L, Keyvan M, Pai G, Mishra R, Haraldsson M, Andresson T, Bragason G, Thosteinsdottir M, Bjornsson JM, Coovert DD, Burghes AH, Gurney ME, Singh J. Thurmond J, et al. J Med Chem. 2008 Feb 14;51(3):449-69. doi: 10.1021/jm061475p. Epub 2008 Jan 19. J Med Chem. 2008. PMID: 18205293 - Regulation of murine survival motor neuron (Smn) protein levels by modifying Smn exon 7 splicing.
DiDonato CJ, Lorson CL, De Repentigny Y, Simard L, Chartrand C, Androphy EJ, Kothary R. DiDonato CJ, et al. Hum Mol Genet. 2001 Nov 1;10(23):2727-36. doi: 10.1093/hmg/10.23.2727. Hum Mol Genet. 2001. PMID: 11726560 - Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients.
Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, Burghes AH. Andreassi C, et al. Hum Mol Genet. 2001 Nov 15;10(24):2841-9. doi: 10.1093/hmg/10.24.2841. Hum Mol Genet. 2001. PMID: 11734549 - Molecular and cellular basis of spinal muscular atrophy.
Jablonka S, Sendtner M. Jablonka S, et al. Amyotroph Lateral Scler Other Motor Neuron Disord. 2003 Sep;4(3):144-9. doi: 10.1080/14660820310011296. Amyotroph Lateral Scler Other Motor Neuron Disord. 2003. PMID: 13129800 Review.
Cited by
- IGF-1R Reduction Triggers Neuroprotective Signaling Pathways in Spinal Muscular Atrophy Mice.
Biondi O, Branchu J, Ben Salah A, Houdebine L, Bertin L, Chali F, Desseille C, Weill L, Sanchez G, Lancelin C, Aïd S, Lopes P, Pariset C, Lécolle S, Côté J, Holzenberger M, Chanoine C, Massaad C, Charbonnier F. Biondi O, et al. J Neurosci. 2015 Aug 26;35(34):12063-79. doi: 10.1523/JNEUROSCI.0608-15.2015. J Neurosci. 2015. PMID: 26311784 Free PMC article. - Phosphatase and tensin homologue: a therapeutic target for SMA.
Godena VK, Ning K. Godena VK, et al. Signal Transduct Target Ther. 2017 Sep 8;2:17038. doi: 10.1038/sigtrans.2017.38. eCollection 2017. Signal Transduct Target Ther. 2017. PMID: 29263925 Free PMC article. Review. - Regulation of Survival Motor Neuron Protein by the Nuclear Factor-Kappa B Pathway in Mouse Spinal Cord Motoneurons.
Arumugam S, Mincheva-Tasheva S, Periyakaruppiah A, de la Fuente S, Soler RM, Garcera A. Arumugam S, et al. Mol Neurobiol. 2018 Jun;55(6):5019-5030. doi: 10.1007/s12035-017-0710-4. Epub 2017 Aug 14. Mol Neurobiol. 2018. PMID: 28808928 - Therapeutics development for spinal muscular atrophy.
Sumner CJ. Sumner CJ. NeuroRx. 2006 Apr;3(2):235-45. doi: 10.1016/j.nurx.2006.01.010. NeuroRx. 2006. PMID: 16554261 Free PMC article. Review. - Defects in pancreatic development and glucose metabolism in SMN-depleted mice independent of canonical spinal muscular atrophy neuromuscular pathology.
Bowerman M, Michalski JP, Beauvais A, Murray LM, DeRepentigny Y, Kothary R. Bowerman M, et al. Hum Mol Genet. 2014 Jul 1;23(13):3432-44. doi: 10.1093/hmg/ddu052. Epub 2014 Feb 4. Hum Mol Genet. 2014. PMID: 24497575 Free PMC article.
References
- Crawford TO, Pardo CA. Neurobiol Dis. 1996;3:97–110. - PubMed
- Munsat TL, Davies KE. Neuromuscul Disord. 1992;2:423–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources