Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory - PubMed (original) (raw)
Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory
Alessia Pascale et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Neuronal ELAV-like proteins (HuB, HuC, and HuD) are highly conserved RNA-binding proteins able to selectively associate with the 3' UTR of a subset of target mRNAs and increase their cytoplasmic stability and rate of translation. We previously demonstrated the involvement of these proteins in learning, reporting that they undergo a sustained up-regulation in the hippocampus of mice trained in a spatial discrimination task. Here, we extend this finding, showing that a similar up-regulation occurs in the hippocampus of rats trained in another spatial learning paradigm, the Morris water maze. HuD, a strictly neuron-specific ELAV-like protein, is shown to increase after learning, with a preferential binding to the cytoskeletal fraction. HuD up-regulation is associated with an enhancement of GAP-43 mRNA and protein levels, with an apparently increased HuD colocalization with the GAP-43 mRNA and an increased association of neuronal ELAV-like proteins with the GAP-43 mRNA. These learning-dependent biochemical events appear to be spatiotemporally controlled, because they do not occur in another brain region involved in learning, the retrosplenial cortex, and at the level of protein expression they show extinction 1 month after training despite memory retention. By contrast, HuD mRNA levels still remain increased after 1 month in the CA1 region. This persistence may have implications for long-term memory recall.
Figures
Fig. 1.
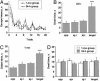
Acquisition and long-term retention of spatial-memory WM task. (A) Similar dynamics of escape latency in the groups of rats trained for biochemical studies of the recent and 1-mo-old spatial memories (n = 10; ref. 10). (B and C) Results of probe tests performed in the same animals 24 h (B) and 1 mo (C) after the last WM training session indicate no decline in spatial memory (n = 5). (D) Absence of spatial preference in both 24-h and 1-mo swimming controls as indicated by rats' dwell time in the pool quadrants (n = 10; ref. 10). Oppquadrant, opposite to training or target quadrant; Aj-r, adjacent right quadrant; Aj-l, adjacent left quadrant. Data represent means ± SE; n = number of tested animals. ****, P < 0.0001, repeated-measures ANOVA with Fisher's post hoc test.
Fig. 2.
Increased expression of the ELAV-like RNA-binding proteins in rats learning a Morris WM spatial task. (A) (Upper) Representative Western blots obtained by using the 16A11 anti-Hu mouse monoclonal 16A11 antibody from the whole-cell lysate or after subcellular fractionation of the hippocampus. (Lower) Mean gray level ratios (means ± SEM) of the determinations of the ELAV-like immunoreactivity measured by Western blots (white bar, SW animals; black bar, TR animals) in the tested hippocampal fractions (n = 6 for each group; *, P < 0.05, **, P < 0.005 Student's t test). Data were normalized to α-tubulin signal. (B) Steady-state levels of HuB, HuC, and HuD mRNAs evaluated by external-standard based real-time quantitative RT-PCR. Measurements obtained from hippocampal total RNA preparations of SW and TR rats were normalized to the signal of GAPDH mRNA and expressed as means ± SEM (n = 6 for each group; **, P < 0.01; ***, P < 0.001 Student's t test).
Fig. 3.
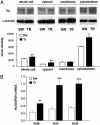
Specific up-regulation of the ELAV-like protein HuD, after Morris WM training, detected at the mRNA and protein levels. (A)(Upper) Representative in situ hybridization results reported for HuD and for a control phylogenetically related gene, BRUNOL3 (BR3), in the hippocampus of SW and TR rats. (Lower) Mean gray levels (±SEM) from autoradiograms of brain sections in the CA1, CA3, and DG+CA4 subfields for six animals in each group (white bar, SW; black bar, TR; *, P < 0.05, Student's t test). (B) Representative Western blots obtained by biochemical fractionation of protein lysates of the hippocampal (HIP) and retrosplenial cortex (RCX) brain regions. α-Tubulin levels for both regions are shown below and were used to normalize the data reported in the text; the statistical analysis is reported in the text.
Fig. 4.
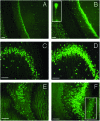
Expression of HuD protein in hippocampal pyramidal, polymorphic (hilar), and granular neurons after Morris WM training in rats. Fluorescence microscopy images showing HuD immunostaining in the CA1 (A and B), CA3 (C and D), and CA4 plus DG (E and F) hippocampal subregions from SW (A, C, and E) and TR (B, D, and F) rats. (Bars = 100 μm.) (B Inset) A pyramidal cell, at higher magnification, in the CA1 area from a TR rat; the staining in the proximal dendritic region is clearly evident. (F Inset) A detail of the granular cell layer in the DG region, at higher magnification, from a TR animal where it is possible to distinguish HuD-positive cells.
Fig. 5.
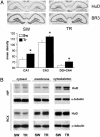
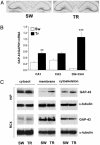
Induction of GAP-43 expression by Morris WM training in the hippocampus, documented as mRNA and protein increase. (A) Representative in situ hybridization showing GAP-43 mRNA distribution in the hippocampus of SW and TR rats. (B) Determination of the steady-state relative levels of GAP-43 mRNA by external-standard based real-time quantitative RT-PCR in the CA1, CA3, and DG+CA4 subfields. Measurements obtained from total hippocampal RNA preparations of SW and TR rats were normalized to the signal of GAPDH mRNA and expressed as means ± SEM (n = six for each group; **, P < 0.005; ***, P < 0.0001, Student's t test). (C) Representative Western blots performed after fractionation of protein lysates of the hippocampal (HIP) and retrosplenial cortex (RCX) brain regions. α-Tubulin levels for both regions were used to normalize the data; the statistical analysis is reported in the text.
Fig. 6.
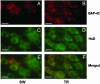
Hippocampal colocalization of GAP-43 mRNA and HuD protein after the Morris WM task. Fluorescence microscopy images taken in the CA4 region of the hippocampus of swimming SW (Left) and TR (Right) animals. (A and B) GAP-43 mRNA signal (red). (C and D) HuD protein signal (green). (E and F) Merged images (the orange color indicates overlapping of the two signals). (Bars = 50 μm.)
Fig. 7.
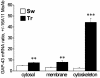
Increased coimmunoprecipitation of ELAV-like proteins and GAP-43 mRNA in the hippocampus after the Morris WM task. The levels of GAP-43 mRNA evaluated by external-standard based real-time quantitative RT-PCR were assessed in the pellet of hippocampal tissue fractions of SW and TR rats immunoprecipitated by the 16A11 MoAb. Values obtained in the presence of the antibody were normalized with the corresponding background values measured in the immunoprecipitation pellet in the absence of any antibody. Data are expressed as means ± SEM (n = 4 for each group; **, P < 0.005; ***, P < 0.001 Student's t test).
Fig. 8.
Increased expression of HuD mRNA in rat hippocampus 1 mo after WM spatial learning. Shown are representative in situ hybridization images reported for HuD in the hippocampus of swimming SW and TR rats. The circled area corresponds to the CA1 regions.
Similar articles
- Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins.
Quattrone A, Pascale A, Nogues X, Zhao W, Gusev P, Pacini A, Alkon DL. Quattrone A, et al. Proc Natl Acad Sci U S A. 2001 Sep 25;98(20):11668-73. doi: 10.1073/pnas.191388398. Proc Natl Acad Sci U S A. 2001. PMID: 11573004 Free PMC article. - In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD.
Bolognani F, Tanner DC, Merhege M, Deschênes-Furry J, Jasmin B, Perrone-Bizzozero NI. Bolognani F, et al. J Neurochem. 2006 Feb;96(3):790-801. doi: 10.1111/j.1471-4159.2005.03607.x. Epub 2006 Jan 9. J Neurochem. 2006. PMID: 16405504 - Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD.
Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Bolognani F, et al. Neurobiol Learn Mem. 2007 May;87(4):635-43. doi: 10.1016/j.nlm.2006.11.004. Epub 2006 Dec 20. Neurobiol Learn Mem. 2007. PMID: 17185008 - Role of HuD and other RNA-binding proteins in neural development and plasticity.
Perrone-Bizzozero N, Bolognani F. Perrone-Bizzozero N, et al. J Neurosci Res. 2002 Apr 15;68(2):121-6. doi: 10.1002/jnr.10175. J Neurosci Res. 2002. PMID: 11948657 Review. - Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction.
Bronicki LM, Jasmin BJ. Bronicki LM, et al. RNA. 2013 Aug;19(8):1019-37. doi: 10.1261/rna.039164.113. RNA. 2013. PMID: 23861535 Free PMC article. Review.
Cited by
- Cytoplasmic RNA-binding proteins and the control of complex brain function.
Darnell JC, Richter JD. Darnell JC, et al. Cold Spring Harb Perspect Biol. 2012 Aug 1;4(8):a012344. doi: 10.1101/cshperspect.a012344. Cold Spring Harb Perspect Biol. 2012. PMID: 22723494 Free PMC article. Review. - Concentration and Localization of Coexpressed ELAV/Hu Proteins Control Specificity of mRNA Processing.
Zaharieva E, Haussmann IU, Bräuer U, Soller M. Zaharieva E, et al. Mol Cell Biol. 2015 Sep;35(18):3104-15. doi: 10.1128/MCB.00473-15. Epub 2015 Jun 29. Mol Cell Biol. 2015. PMID: 26124284 Free PMC article. - Intra-axonal translation of Khsrp mRNA slows axon regeneration by destabilizing localized mRNAs.
Patel P, Buchanan CN, Zdradzinski MD, Sahoo PK, Kar AN, Lee SJ, Vaughn LS, Urisman A, Oses-Prieto J, Dell'Orco M, Cassidy DE, Costa ID, Miller S, Thames E, Smith TP, Burlingame AL, Perrone-Bizzozero N, Twiss JL. Patel P, et al. Nucleic Acids Res. 2022 Jun 10;50(10):5772-5792. doi: 10.1093/nar/gkac337. Nucleic Acids Res. 2022. PMID: 35556128 Free PMC article. - Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function.
Gardiner AS, Twiss JL, Perrone-Bizzozero NI. Gardiner AS, et al. Biomolecules. 2015 Oct 23;5(4):2903-18. doi: 10.3390/biom5042903. Biomolecules. 2015. PMID: 26512708 Free PMC article. Review. - Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA.
Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Zhu H, et al. Mol Cell Biol. 2008 Feb;28(4):1240-51. doi: 10.1128/MCB.01509-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086893 Free PMC article.
References
- Robinow, S. & White, K. (1991) J. Neurobiol. 22, 443–461. - PubMed
- Wakamatsu, Y. & Weston, J. A. (1997) Development (Cambridge, U.K.) 124, 3449–3460. - PubMed
- Robinow, S., Campos, A. R., Yao, K. M. & White, K. (1988) Science 242, 1570–1572. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous