Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer - PubMed (original) (raw)
Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer
Nasser Tahbaz et al. EMBO Rep. 2004 Feb.
Abstract
PAZ PIWI domain (PPD) proteins, together with the RNA cleavage products of Dicer, form ribonucleoprotein complexes called RNA-induced silencing complexes (RISCs). RISCs mediate gene silencing through targeted messenger RNA cleavage and translational suppression. The PAZ domains of PPD and Dicer proteins were originally thought to mediate binding between PPD proteins and Dicer, although no evidence exists to support this theory. Here we show that PAZ domains are not required for PPD protein-Dicer interactions. Rather, a subregion of the PIWI domain in PPD proteins, the PIWI-box, binds directly to the Dicer RNase III domain. Stable binding between PPD proteins and Dicer was dependent on the activity of Hsp90. Unexpectedly, binding of PPD proteins to Dicer inhibits the RNase activity of this enzyme in vitro. Lastly, we show that PPD proteins and Dicer are present in soluble and membrane-associated fractions, indicating that interactions between these two types of proteins may occur in multiple compartments.
Figures
Figure 1
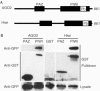
PIWI domains interact with Dicer. (A) Schematic representation of AGO2 and Hiwi GST-fusion proteins used for binding studies. PIWI domains, PIWI-box (Pb) and PAZ domains are indicated. (B) GST and GST–PPD fusion proteins were co-expressed with CFP–Dicer in HEK293T cells. Proteins were purified on glutathione–sepharose beads, resolved by SDS–PAGE, and visualized by immunoblotting with antibodies to GST and GFP.
Figure 2
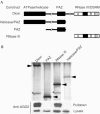
The Dicer RNase III domain is required for binding to AGO2. (A) Schematic representation of Dicer and GST-fusion proteins used for binding studies. The three regions of Dicer are indicated: ATPase/helicase, PAZ and RNase III domains. The RNase III domain is further subdivided to indicate the presence of two RNase III motifs A and B and a dsRNA-binding domain (DSRM; D). (B) Plasmids encoding GST or GST fused to domains of Dicer were transfected into a cell line overexpressing AGO2. Following GST pulldowns, proteins were visualized by silver staining (top panel) and immunoblotting (bottom panels). Arrowheads point to the silverstained GST-fusion proteins, whereas AGO2 is indicated by asterisks.
Figure 3
PPD–Dicer interactions are direct. (A) Protein A–sepharose beads coated with affinity-purified Abs against Dicer or the control protein GAR1 were incubated with mixtures of purified Dicer and GST–AGO2 (upper panel), GST–Hiwi (middle panel) or GST alone (lower panel). Bead-associated (B) and unbound (S) material was subjected to SDS–PAGE and immunoblotting with anti-GST Abs. (B) Different combinations of pGBKT7 (AGO2 or Hiwi subdomains) and pGADT7 (Dicer subdomains) constructs were transformed into AH109 strain and tested for interaction by growth on selective media. Four serial dilutions were spotted for each combination. Scoring ranged from − to ++++ depending upon how well the strains grew on media lacking leucine, tryptophan, adenine and histidine. Graded examples of some PPD–Dicer interactions are shown. A complete listing of all the tested interactions is shown in supplementary table 2 online.
Figure 4
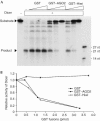
PPD proteins inhibit the RNase activity of Dicer in vitro. (A) Effect of Hiwi and AGO2 on Dicer-mediated cleavage of dsRNA. Increasing amounts (indicated in B) of GST–Hiwi, GST–AGO2 or GST alone were added to the cleavage reactions containing 130-bp 32P-labelled dsRNA substrate. Reaction products were analysed by 8 M urea PAGE. The positions of 14-, 21- and 27-nt size markers are indicated (the 21-nt marker was only visible upon longer exposure). (B) PhosphorImager quantitation of the cleavage reactions from a typical experiment.
Figure 5
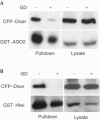
GD inhibits the binding of PPD proteins to Dicer. GST–AGO2 (A) or GST–Hiwi (B) were transiently co-expressed with CFP–Dicer in HEK293T cells. Cells were incubated with (+) or without (−) 5 μM GD for 2 h before lysis. Proteins were recovered on glutathione–sepharose beads and visualized by immunoblotting. CFP–Dicer was detected using a polyclonal anti-GFP Ab, whereas GST–AGO2 and GST–Hiwi were detected using anti-GST Abs.
Figure 6
Dicer and PPD proteins are present in soluble and membrane-associated pools. Homogenates of HEK293T cells transiently expressing GST–Hiwi were subjected to membrane flotation assays on discontinuous sucrose gradients. Fractions were collected from the tops (T) of gradients, TCA precipitated, resolved by SDS–PAGE and subjected to immunoblotting with Abs specific for AGO2, GST, Dicer, calnexin and Hsp90. Membrane-associated material is found at the top of the gradient, whereas soluble proteins remain in the bottom (B) three fractions.
Similar articles
- The role of PACT in the RNA silencing pathway.
Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. Lee Y, et al. EMBO J. 2006 Feb 8;25(3):522-32. doi: 10.1038/sj.emboj.7600942. Epub 2006 Jan 19. EMBO J. 2006. PMID: 16424907 Free PMC article. - Human dicer: purification, properties, and interaction with PAZ PIWI domain proteins.
Kolb FA, Zhang H, Jaronczyk K, Tahbaz N, Hobman TC, Filipowicz W. Kolb FA, et al. Methods Enzymol. 2005;392:316-36. doi: 10.1016/S0076-6879(04)92019-8. Methods Enzymol. 2005. PMID: 15644190 - Unknown Areas of Activity of Human Ribonuclease Dicer: A Putative Deoxyribonuclease Activity.
Wojnicka M, Szczepanska A, Kurzynska-Kokorniak A. Wojnicka M, et al. Molecules. 2020 Mar 20;25(6):1414. doi: 10.3390/molecules25061414. Molecules. 2020. PMID: 32244942 Free PMC article. - Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases.
Ciechanowska K, Pokornowska M, Kurzyńska-Kokorniak A. Ciechanowska K, et al. Int J Mol Sci. 2021 Jan 9;22(2):616. doi: 10.3390/ijms22020616. Int J Mol Sci. 2021. PMID: 33435485 Free PMC article. Review. - The mechanism of RNase III action: how dicer dices.
Ji X. Ji X. Curr Top Microbiol Immunol. 2008;320:99-116. doi: 10.1007/978-3-540-75157-1_5. Curr Top Microbiol Immunol. 2008. PMID: 18268841 Review.
Cited by
- RNA interference in infectious tropical diseases.
Kang S, Hong YS. Kang S, et al. Korean J Parasitol. 2008 Mar;46(1):1-15. doi: 10.3347/kjp.2008.46.1.1. Korean J Parasitol. 2008. PMID: 18344671 Free PMC article. Review. - Molecular mechanisms of RNA interference.
Wilson RC, Doudna JA. Wilson RC, et al. Annu Rev Biophys. 2013;42:217-39. doi: 10.1146/annurev-biophys-083012-130404. Annu Rev Biophys. 2013. PMID: 23654304 Free PMC article. Review. - Comparative analysis of the Dicer-like gene family reveals loss of miR162 target site in SmDCL1 from Salvia miltiorrhiza.
Shao F, Qiu D, Lu S. Shao F, et al. Sci Rep. 2015 May 13;5:9891. doi: 10.1038/srep09891. Sci Rep. 2015. PMID: 25970825 Free PMC article. - The Argonaute protein family.
Höck J, Meister G. Höck J, et al. Genome Biol. 2008;9(2):210. doi: 10.1186/gb-2008-9-2-210. Epub 2008 Feb 26. Genome Biol. 2008. PMID: 18304383 Free PMC article. Review. - Control of the localization and function of a miRNA silencing component TNRC6A by Argonaute protein.
Nishi K, Takahashi T, Suzawa M, Miyakawa T, Nagasawa T, Ming Y, Tanokura M, Ui-Tei K. Nishi K, et al. Nucleic Acids Res. 2015 Nov 16;43(20):9856-73. doi: 10.1093/nar/gkv1026. Epub 2015 Oct 7. Nucleic Acids Res. 2015. PMID: 26446993 Free PMC article.
References
- Baulcombe D (2001) RNA silencing: diced defence. Nature 409: 295–296 - PubMed
- Bernstein E, Caudy A, Hammond S, Hannon G (2001) Role for bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 - PubMed
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases