Increasing the power and efficiency of disease-marker case-control association studies through use of allele-sharing information - PubMed (original) (raw)
Comparative Study
doi: 10.1086/381652. Epub 2004 Feb 2.
Affiliations
- PMID: 14752704
- PMCID: PMC1182257
- DOI: 10.1086/381652
Comparative Study
Increasing the power and efficiency of disease-marker case-control association studies through use of allele-sharing information
Tasha E Fingerlin et al. Am J Hum Genet. 2004 Mar.
Abstract
Case-control disease-marker association studies are often used in the search for variants that predispose to complex diseases. One approach to increasing the power of these studies is to enrich the case sample for individuals likely to be affected because of genetic factors. In this article, we compare three case-selection strategies that use allele-sharing information with the standard strategy that selects a single individual from each family at random. In affected sibship samples, we show that, by carefully selecting sibships and/or individuals on the basis of allele sharing, we can increase the frequency of disease-associated alleles in the case sample. When these cases are compared with unrelated controls, the difference in the frequency of the disease-associated allele is therefore also increased. We find that, by choosing the affected sib who shows the most evidence for pairwise allele sharing with the other affected sibs in families, the test statistic is increased by >20%, on average, for additive models with modest genotype relative risks. In addition, we find that the per-genotype information associated with the allele sharing-based strategies is increased compared with that associated with random selection of a sib for genotyping. Even though we select sibs on the basis of a nonparametric statistic, the additional gain for selection based on the unknown underlying mode of inheritance is minimal. We show that these properties hold even when the power to detect linkage to a region in the entire sample is negligible. This approach can be extended to more-general pedigree structures and quantitative traits.
Figures
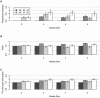
Figure 1
Additive models with GRR2 ⩽3, mean ± 2 SD. A, Proportional increase in disease-allele frequency difference for sharing-based strategies compared with the AR strategy. B, Ratio of test statistics for sharing-based strategies to test statistic for the AR strategy. C, Ratio of per-genotype information for sharing-based strategies to per-genotype information for the AR strategy.
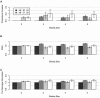
Figure A
Dominant models with GRR2 ⩽3, mean ± 2 SD. A, Proportional increase in disease-allele frequency difference for sharing-based strategies compared with the AR strategy. B, Ratio of test statistics for sharing-based strategies to test statistic for the AR strategy. C, Ratio of per-genotype information for sharing-based strategies to per-genotype information for the AR strategy.
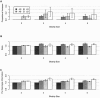
Figure B
Recessive models with GRR2 ⩽3, mean ± 2 SD. A, Proportional increase in disease-allele frequency difference for sharing-based strategies compared with the AR strategy. B, Ratio of test statistics for sharing-based strategies to test statistic for the AR strategy. C, Ratio of per-genotype information for sharing-based strategies to per-genotype information for the AR strategy.
Similar articles
- Use of parents, sibs, and unrelated controls for detection of associations between genetic markers and disease.
Schaid DJ, Rowland C. Schaid DJ, et al. Am J Hum Genet. 1998 Nov;63(5):1492-506. doi: 10.1086/302094. Am J Hum Genet. 1998. PMID: 9792877 Free PMC article. - Assessing whether an allele can account in part for a linkage signal: the Genotype-IBD Sharing Test (GIST).
Li C, Scott LJ, Boehnke M. Li C, et al. Am J Hum Genet. 2004 Mar;74(3):418-31. doi: 10.1086/381712. Epub 2004 Feb 6. Am J Hum Genet. 2004. PMID: 14872409 Free PMC article. - The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases I. DNA pooling.
Risch N, Teng J. Risch N, et al. Genome Res. 1998 Dec;8(12):1273-88. doi: 10.1101/gr.8.12.1273. Genome Res. 1998. PMID: 9872982 Review. - Sibship T2 association tests of complex diseases for tightly linked markers.
Fan R, Knapp M. Fan R, et al. Hum Genomics. 2005 Jun;2(2):90-112. doi: 10.1186/1479-7364-2-2-90. Hum Genomics. 2005. PMID: 16004725 Free PMC article. Review.
Cited by
- Sampling strategies for rare variant tests in case-control studies.
Zöllner S. Zöllner S. Eur J Hum Genet. 2012 Oct;20(10):1085-91. doi: 10.1038/ejhg.2012.58. Epub 2012 Apr 18. Eur J Hum Genet. 2012. PMID: 22510851 Free PMC article. - Family-based association tests for genomewide association scans.
Chen WM, Abecasis GR. Chen WM, et al. Am J Hum Genet. 2007 Nov;81(5):913-26. doi: 10.1086/521580. Epub 2007 Sep 18. Am J Hum Genet. 2007. PMID: 17924335 Free PMC article. - Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26.
Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, Carey A, Ewen-White KR, Duffy DL, O'connor DT, Barlow DH, Martin NG, Kennedy SH. Treloar SA, et al. Am J Hum Genet. 2005 Sep;77(3):365-76. doi: 10.1086/432960. Epub 2005 Jul 21. Am J Hum Genet. 2005. PMID: 16080113 Free PMC article. - A powerful strategy to account for multiple testing in the context of haplotype analysis.
Becker T, Knapp M. Becker T, et al. Am J Hum Genet. 2004 Oct;75(4):561-70. doi: 10.1086/424390. Epub 2004 Jul 30. Am J Hum Genet. 2004. PMID: 15290652 Free PMC article. - Family-based samples can play an important role in genetic association studies.
Lange EM, Sun J, Lange LA, Zheng SL, Duggan D, Carpten JD, Gronberg H, Isaacs WB, Xu J, Chang BL. Lange EM, et al. Cancer Epidemiol Biomarkers Prev. 2008 Sep;17(9):2208-14. doi: 10.1158/1055-9965.EPI-08-0183. Cancer Epidemiol Biomarkers Prev. 2008. PMID: 18768484 Free PMC article.
References
Electronic-Database Information
- Merlin Web site, http://www.sph.umich.edu/csg/abecasis/Merlin/ (for Merlin version 0.9.15 and above, in which the method used for the selection of informative cases is implemented)
References
Publication types
MeSH terms
Substances
Grants and funding
- Z01 HG000040/ImNIH/Intramural NIH HHS/United States
- R01 HG002651/HG/NHGRI NIH HHS/United States
- R01 HG000376/HG/NHGRI NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- HG02651/HG/NHGRI NIH HHS/United States
- HG00376/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical