Oxidative protein folding in eukaryotes: mechanisms and consequences - PubMed (original) (raw)
Review
Oxidative protein folding in eukaryotes: mechanisms and consequences
Benjamin P Tu et al. J Cell Biol. 2004.
Abstract
The endoplasmic reticulum (ER) provides an environment that is highly optimized for oxidative protein folding. Rather than relying on small molecule oxidants like glutathione, it is now clear that disulfide formation is driven by a protein relay involving Ero1, a novel conserved FAD-dependent enzyme, and protein disulfide isomerase (PDI); Ero1 is oxidized by molecular oxygen and in turn acts as a specific oxidant of PDI, which then directly oxidizes disulfide bonds in folding proteins. While providing a robust driving force for disulfide formation, the use of molecular oxygen as the terminal electron acceptor can lead to oxidative stress through the production of reactive oxygen species and oxidized glutathione. How Ero1p distinguishes between the many different PDI-related proteins and how the cell minimizes the effects of oxidative damage from Ero1 remain important open questions.
Copyright The Rockefeller University Press
Figures
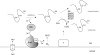
Figure 1.
Schematic model of oxidative protein folding in the yeast ER. The formation of disulfide bonds in the ER is driven by Ero1p. FAD-bound Ero1p oxidizes PDI, which then subsequently oxidizes folding proteins directly. FAD-bound Ero1p then passes electrons to molecular oxygen, perhaps resulting in the production of ROS. FAD, which is synthesized in the cytosol, can readily enter the ER lumen and stimulate the activity of Ero1p. Disulfide isomerization and reduction may be performed by some of the four homologues of PDI, Eug1p, Mpd1p, Mpd2p, or Eps1p, in addition to PDI itself. Reduced glutathione (GSH) may also assist in disulfide reduction, resulting in the production of oxidized glutathione (GSSG).
Similar articles
- Biochemical basis of oxidative protein folding in the endoplasmic reticulum.
Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Tu BP, et al. Science. 2000 Nov 24;290(5496):1571-4. doi: 10.1126/science.290.5496.1571. Science. 2000. PMID: 11090354 - Role of the ERO1-PDI interaction in oxidative protein folding and disease.
Shergalis AG, Hu S, Bankhead A 3rd, Neamati N. Shergalis AG, et al. Pharmacol Ther. 2020 Jun;210:107525. doi: 10.1016/j.pharmthera.2020.107525. Epub 2020 Mar 20. Pharmacol Ther. 2020. PMID: 32201313 Free PMC article. Review. - Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum.
Molteni SN, Fassio A, Ciriolo MR, Filomeni G, Pasqualetto E, Fagioli C, Sitia R. Molteni SN, et al. J Biol Chem. 2004 Jul 30;279(31):32667-73. doi: 10.1074/jbc.M404992200. Epub 2004 May 25. J Biol Chem. 2004. PMID: 15161913 - Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum.
Vitu E, Kim S, Sevier CS, Lutzky O, Heldman N, Bentzur M, Unger T, Yona M, Kaiser CA, Fass D. Vitu E, et al. J Biol Chem. 2010 Jun 11;285(24):18155-65. doi: 10.1074/jbc.M109.064931. Epub 2010 Mar 26. J Biol Chem. 2010. PMID: 20348090 Free PMC article. - Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum.
Hudson DA, Gannon SA, Thorpe C. Hudson DA, et al. Free Radic Biol Med. 2015 Mar;80:171-82. doi: 10.1016/j.freeradbiomed.2014.07.037. Epub 2014 Aug 1. Free Radic Biol Med. 2015. PMID: 25091901 Free PMC article. Review.
Cited by
- Effects of Apoptin-Induced Endoplasmic Reticulum Stress on Lipid Metabolism, Migration, and Invasion of HepG-2 Cells.
Zhu Y, Li Y, Bai B, Shang C, Fang J, Cong J, Li W, Li S, Song G, Liu Z, Zhao J, Li X, Zhu G, Jin N. Zhu Y, et al. Front Oncol. 2021 Feb 26;11:614082. doi: 10.3389/fonc.2021.614082. eCollection 2021. Front Oncol. 2021. PMID: 33718168 Free PMC article. - The unfolded protein response in immunity and inflammation.
Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. Grootjans J, et al. Nat Rev Immunol. 2016 Aug;16(8):469-84. doi: 10.1038/nri.2016.62. Epub 2016 Jun 27. Nat Rev Immunol. 2016. PMID: 27346803 Free PMC article. Review. - Regulation of antioxidants in cancer.
Hecht F, Zocchi M, Alimohammadi F, Harris IS. Hecht F, et al. Mol Cell. 2024 Jan 4;84(1):23-33. doi: 10.1016/j.molcel.2023.11.001. Epub 2023 Nov 28. Mol Cell. 2024. PMID: 38029751 Free PMC article. Review. - NPGPx (GPx7): a novel oxidative stress sensor/transmitter with multiple roles in redox homeostasis.
Chen YI, Wei PC, Hsu JL, Su FY, Lee WH. Chen YI, et al. Am J Transl Res. 2016 Apr 15;8(4):1626-40. eCollection 2016. Am J Transl Res. 2016. PMID: 27186289 Free PMC article. Review. - The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities.
Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, Smith SB, Ganapathy V, Maher P. Lewerenz J, et al. Antioxid Redox Signal. 2013 Feb 10;18(5):522-55. doi: 10.1089/ars.2011.4391. Epub 2012 Aug 3. Antioxid Redox Signal. 2013. PMID: 22667998 Free PMC article. Review.
References
- Bader, M., W. Muse, D.P. Ballou, C. Gassner, and J.C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell. 98:217–227. - PubMed
- Bader, M.W., T. Xie, C.A. Yu, and J.C. Bardwell. 2000. Disulfide bonds are generated by quinone reduction. J. Biol. Chem. 275:26082–26088. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases