Fission yeast Arp6 is required for telomere silencing, but functions independently of Swi6 - PubMed (original) (raw)
Fission yeast Arp6 is required for telomere silencing, but functions independently of Swi6
Masaru Ueno et al. Nucleic Acids Res. 2004.
Abstract
The actin-related proteins (Arps), which are subdivided into at least eight subfamilies, are conserved from yeast to humans. A member of the Arp6 subfamily in Drosophila, Arp4/Arp6, co-localizes with heterochromatin protein 1 (HP1) in pericentric heterochromatin. Fission yeast Schizosaccharomyces pombe possesses both an HP1 homolog and an Arp6 homolog. However, the function of S.pombe Arp6 has not been characterized yet. We found that deletion of arp6(+) impaired telomere silencing, but did not affect centromere silencing. Chromatin immunoprecipitation assays revealed that Arp6 bound to the telomere region. However, unlike Drosophila Arp4/Arp6, S.pombe Arp6 was distributed throughout nuclei. The binding of Arp6 to telomere DNA was not affected by deletion of swi6(+). Moreover, the binding of Swi6 to telomere ends was not affected by deletion of arp6(+). These results suggest that Arp6 and Swi6 function independently at telomere ends. We propose that the Arp6-mediated repression mechanism works side by side with Swi6-based telomere silencing in S.pombe.
Figures
Figure 1
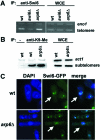
Deletion of arp6+ affects telomere silencing, but not centromere silencing. (A) Serial dilution assay on no histidine (SD-histidine) and non-selective (YPAD) plates. Telomere silencing in wild-type (FY1862), _arp6_Δ (TM001) and _taz1_Δ (KT21-UHA) cells was studied. (B) Quantitative RT–PCR was performed on RNA prepared from wild-type (FY1862) and _arp6_Δ cells (TM001) using primers to amplify His3 mRNA and to amplify Eno1 mRNA as a control. The RT–PCR products, His3 cDNA (his3) and Eno1 cDNA (eno1), separated by agarose gel are shown. (C) Serial dilution assay on low adenine and non-selective (YPAD) plates. Centromere silencing in wild-type cells (FY1862), _arp6_Δ (TM001) and _swi6_Δ (TK017) was studied.
Figure 2
Arp6-Myc is bound to telomere DNA in ChIP assay. Untagged wild-type control (JY746), Arp6-Myc (TM002), Arp6-Myc _swi6_Δ (TM004) and Arp6-Myc _taz1_Δ (TM005) cells were used. PCR was performed on whole-cell extracts (WCE) and on chromatin immunoprecipitates (IP: anti-Myc) using primers to amplify telomere DNA (telomere) and primers to amplify DNA from the eno1+ gene (eno1).
Figure 3
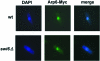
Arp6 is dispensable for localization of Swi6 to heterochromatin. (A) The binding of Swi6 to telomeres in _arp6_Δ cells was studied by the ChIP assay. Wild-type (JY746), _swi6_Δ (PY614) and _arp6_Δ (TM001) cells were used. PCR was performed on whole-cell extracts (WCE) and on chromatin immunoprecipitates (IP: anti-Swi6) using primers to amplify telomere DNA (telomere) and primers to amplify DNA from the eno1+ gene (eno1). (B) H3 Lys9 methylation at subtelomere region (∼2 kb away from telomere ends) was studied by the ChIP assay. Wild-type (JY746) and _arp6_Δ (TM001) cells were used. PCR was performed on whole-cell extracts (WCE) and on chromatin immunoprecipitates (with anti-dimethyl-histone H3-K9 antibody) (IP: anti-K9-Me) using primers to amplify subtelomere DNA (subtelomere) and primers to amplify DNA from the act1+ gene (act1). For control experiment (IP: –), extract of wild-type cells was immunoprecipitated without anti-dimethyl-histone H3-K9 antibody. (C) Localization of Swi6-GFP to silenced chromosomal locations was not affected by deletion of arp6+. Cells expressing swi6+-GFP in the wild-type strain (PY612) or in _arp6_Δ cells (TM006) were fixed with methanol and stained with DAPI. The positions of Swi6-GFP are indicated by arrows.
Figure 4
Schizosaccharomyces pombe Arp6 is distributed throughout nuclei. Indirect immunofluorescence microscopy of Arp6-Myc (wt, TM002) and Arp6-Myc _swi6_Δ cells (_swi6_Δ, TM004). Anti-Myc antibody was used to detect Myc-tagged Arp6.
Similar articles
- Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres.
Kanoh J, Sadaie M, Urano T, Ishikawa F. Kanoh J, et al. Curr Biol. 2005 Oct 25;15(20):1808-19. doi: 10.1016/j.cub.2005.09.041. Curr Biol. 2005. PMID: 16243027 - Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1.
Ohfuchi E, Kato M, Sasaki M, Sugimoto K, Oma Y, Harata M. Ohfuchi E, et al. Eur J Cell Biol. 2006 May;85(5):411-21. doi: 10.1016/j.ejcb.2005.12.006. Epub 2006 Feb 17. Eur J Cell Biol. 2006. PMID: 16487625 - A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast.
Sadaie M, Iida T, Urano T, Nakayama J. Sadaie M, et al. EMBO J. 2004 Oct 1;23(19):3825-35. doi: 10.1038/sj.emboj.7600401. Epub 2004 Sep 16. EMBO J. 2004. PMID: 15372076 Free PMC article. - Destabilizing heterochromatin: Does Swi6/HP1 make the choice?
Verdel A. Verdel A. Mol Cell. 2006 Jun 23;22(6):709-710. doi: 10.1016/j.molcel.2006.06.004. Mol Cell. 2006. PMID: 16793539 Review. - Studies on the mechanism of RNAi-dependent heterochromatin assembly.
Moazed D, Bühler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villén J, Gygi SP. Moazed D, et al. Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
- Meiotic telomere clustering requires actin for its formation and cohesin for its resolution.
Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Trelles-Sticken E, et al. J Cell Biol. 2005 Jul 18;170(2):213-23. doi: 10.1083/jcb.200501042. J Cell Biol. 2005. PMID: 16027219 Free PMC article. - Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1.
Hansen KR, Ibarra PT, Thon G. Hansen KR, et al. Nucleic Acids Res. 2006 Jan 10;34(1):78-88. doi: 10.1093/nar/gkj415. Print 2006. Nucleic Acids Res. 2006. PMID: 16407326 Free PMC article. - The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis.
Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. Greaves IK, et al. Mol Cell Biol. 2006 Jul;26(14):5394-405. doi: 10.1128/MCB.00519-06. Mol Cell Biol. 2006. PMID: 16809775 Free PMC article. - Chapter 5. Nuclear actin-related proteins in epigenetic control.
Meagher RB, Kandasamy MK, McKinney EC, Roy E. Meagher RB, et al. Int Rev Cell Mol Biol. 2009;277:157-215. doi: 10.1016/S1937-6448(09)77005-4. Int Rev Cell Mol Biol. 2009. PMID: 19766970 Free PMC article. Review. - SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis.
Choi K, Kim S, Kim SY, Kim M, Hyun Y, Lee H, Choe S, Kim SG, Michaels S, Lee I. Choi K, et al. Plant Cell. 2005 Oct;17(10):2647-60. doi: 10.1105/tpc.105.035485. Epub 2005 Sep 9. Plant Cell. 2005. PMID: 16155178 Free PMC article.
References
- Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. - PubMed
- Eissenberg J.C. and Elgin,S.C. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev., 10, 204–210. - PubMed
- Weiler K.S. and Wakimoto,B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. - PubMed
- Lorentz A., Ostermann,K., Fleck,O. and Schmidt,H. (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene, 143, 139–143. - PubMed
- Allshire R.C., Nimmo,E.R., Ekwall,K., Javerzat,J.P. and Cranston,G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev., 9, 218–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials