An unappreciated role for RNA surveillance - PubMed (original) (raw)
An unappreciated role for RNA surveillance
R Tyler Hillman et al. Genome Biol. 2004.
Abstract
Background: Nonsense-mediated mRNA decay (NMD) is a eukaryotic mRNA surveillance mechanism that detects and degrades mRNAs with premature termination codons (PTC+ mRNAs). In mammals, a termination codon is recognized as premature if it lies more than about 50 nucleotides upstream of the final intron position. More than a third of reliably inferred alternative splicing events in humans have been shown to result in PTC+ mRNA isoforms. As the mechanistic details of NMD have only recently been elucidated, we hypothesized that many PTC+ isoforms may have been cloned, characterized and deposited in the public databases, even though they would be targeted for degradation in vivo.
Results: We analyzed the human alternative protein isoforms described in the SWISS-PROT database and found that 144 (5.8% of 2,483) isoform sequences amenable to analysis, from 107 (7.9% of 1,363) SWISS-PROT entries, derive from PTC+ mRNA.
Conclusions: For several of the PTC+ isoforms we identified, existing experimental evidence can be reinterpreted and is consistent with the action of NMD to degrade the transcripts. Several genes with mRNA isoforms that we identified as PTC+--calpain-10, the CDC-like kinases (CLKs) and LARD--show how previous experimental results may be understood in light of NMD.
Figures
Figure 1
Recognition of premature termination codons in humans is splicing dependent. (a) During pre-mRNA processing, introns are removed and a set of proteins called the exon-junction complex is deposited. According to the current model for mammalian NMD, these complexes serve to facilitate transport from the nucleus and to remember the gene structure. During the first, pioneering, round of translation, the ribosome will displace all exon-junction complexes in its path until it reaches a stop codon. If the termination codon is on or near the final exon, as is the case for most genes, the ribosome will have displaced all exon-junction complexes. The mRNA will then undergo multiple rounds of translation. (b) If the termination codon is sufficiently far upstream of the final intron position, exon-junction complexes will remain. Interactions ensue that result in the degradation of the mRNA by NMD.
Figure 2
Many human alternative isoforms in SWISS-PROT derive from PTC+ mRNAs. (a) We analyzed each of the human SWISS-PROT entries containing a VARSPLIC line in its feature table, using this information to assemble protein isoform sequences. Ambiguous VARSPLIC entries led us to discard five entries from our analysis at this point. (b) We next identified cDNA/mRNA sequences corresponding to each protein isoform assembled from SWISS-PROT. BLAST was used to align each protein isoform sequence to translated cDNA/mRNA sequences in GenBank and Refseq, filtering to ensure only high confidence matches. To obtain the coding sequence of each mRNA/cDNA sequence, we used LocusLink to map each to the correct human genomic contig sequence from the NCBI human genome build 30. We referred to the CDS feature of each GenBank or RefSeq cDNA/mRNA record to identify stop codon locations. (c) We used the SPIDEY mRNA-to-genomic DNA alignment program to determine the gene structure of each mRNA/cDNA isoform sequence. After generating these gene structures, we could determine the PTC+ status on the basis of stop codon location relative to exon-exon junctions. If the termination codon was found to be more than 50 nucleotides upstream of the final intron, the transcript was deemed PTC+ and designated a candidate target of NMD according to the model of mammalian PTC recognition. (d) Each putative PTC+ isoform was manually inspected for errors in gene structure prediction. These errors include false exon predictions due to poly(A) tails and cDNA/mRNA sequence not seen in the corresponding genomic sequence.
Figure 3
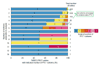
SWISS-PROT entries with multiple isoforms amenable to analysis generate more PTC+ isoforms. We categorized SWISS-PROT entries by the number of isoforms that are amenable to our analysis, and we then determined how many contained a PTC. Each bar shows the number of PTC+ isoforms generated for all SWISS-PROT entries that had the indicated number of isoforms amenable to analysis. Bar components indicate how many entries had a given number of PTC+ isoforms. For example, the bar labeled '3' contains data for the 113 SWISS-PROT entries that had 3 isoforms amenable to analysis. 86% of these had no PTC+ isoforms, 10% had one PTC+ isoform, and 4% had 2 PTC+ isoforms. The bar components outlined in green were SWISS-PROT entries for which all amenable isoforms had a PTC. Entries with multiple isoforms amenable to analysis were more likely to produce at least one PTC+ isoform. This study only considered entries with at least two isoforms in the SWISS-PROT database. For many entries only a single isoform is amenable to analysis, however.
Figure 4
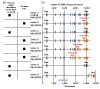
Published expression levels of calpain-10 isoforms are consistent with NMD prediction. (a) A report from Horikawa and co-workers [43] found eight alternative isoforms of calpain-10, of which four are expressed in low abundance. Our analysis found this exact set of four low-abundance isoforms to contain PTCs. (b) Gene structures of alternative mRNA isoforms of calpain-10 show the patterns of alternative splicing and indicate locations of PTCs. Also shown is the position of UCSNP-43, an intronic polymorphism that has been statistically linked to type II diabetes susceptibility in a variety of populations.
Figure 5
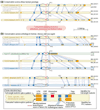
Splicing to generate a premature termination codon is evolutionarily conserved in CLKs. The CDC-like kinases (CLKs) are splicing regulators that affect splicing decisions through the phosphorylation of SR proteins. (a) Our screen of SWISS-PROT revealed that human CLK1, CLK2 and CLK3 paralogs all generate PTC+ alternative isoforms. The splicing pattern that generates these isoforms, skipping exon 4, is conserved in each. This splicing pattern causes a frameshift and a PTC. The percent identities from global alignments between corresponding exons and introns are shown in purple. (b) CLKs were identified in mouse through existing annotation and in the predicted genes of the sea squirt C. intestinalis using an HMM constructed with annotated CLKs from a variety of organisms. An EST analysis revealed that the alternative splicing pattern that generates PTC+ alternative isoforms was conserved in all three sets of orthologs in human and mouse. The same splicing pattern was also found in the only C. intestinalis homolog. A relatively high degree of sequence similarity was found to be present in the introns flanking the alternative exon.
Figure 6
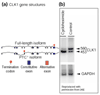
Cycloheximide increases abundance of CLK1 PTC+ isoform. (a) Gene structures of CLK1 full-length and PTC+ isoforms as determined by our analysis. (b) Menegay et al. [48] performed the RT-PCR analysis of CLK1 isoforms; Figure 8 of that analysis [48] is reproduced here with permission (© Company of Biologists Ltd.). The 560 bp fragment corresponds to the full-length CLK1 isoform; the 453 bp fragment corresponds to the PTC+ CLK1 isoform. The analysis shows that cycloheximide, but not UV irradiation or high salt (data not shown), increased the relative abundance of the CLK1 isoform containing a premature termination codon. As cycloheximide is a potent inhibiter of NMD (see, for example, [28,51-53]), this result suggests that the CLK1 PTC+ isoform is degraded by NMD. Menegay et al. [48] describe their figure as follows: "Shift in PCR products of splice forms with cycloheximide. Control or PC12 cells treated with 10 μg/ml cycloheximide for 60 minutes were harvested, RNA was extracted, and RT-PCR was performed. [...] PCR products of the 560 bp full-length form or the 453 kinase-less form of CLK1 message shown. [...] PCR of GAPDH controls from each sample to control for RNA loading."
Figure 7
LARD/TNFRSF12/DR3/Apo3 expression correlates with PTC+ status. LARD is an alternatively spliced death-domain-containing member of the tumor necrosis factor receptor family (TNFR). However, only the major splice variant (isoform 1) contains the death domain and is capable of inducing apoptosis. The splicing distribution of LARD isoforms has been shown to change on lymphocyte activation, suggesting that alternative splicing may be a control point regulating lymphocyte proliferation [55]. (a) Screaton et al. [55] showed that, before lymphocyte activation, only LARD isoforms 2, 3, 4, 5 and 6 are expressed. Primary blood lymphocytes treated with an activating agent were found instead to express the major, apoptosis-promoting splice variant (isoform 1) almost exclusively. This panel is reproduced with permission from Figure 6a of [55] (© National Academy of Sciences). Screaton et al. [55] describe their figure as follows: "Southern blots of reverse transcriptase-PCR of LARD cDNA with primers F LARD Kpn and R LARD Xba probed with 32P-labeled primer F LARD Xba. Lanes: 1, CD4+ cells; 2, CD8+ cells; 3, B cells; 4 PHA-blasted PBL; 5, negative control." (b) LARD isoforms 2, 3, 4, 5 and 6 were found in our analysis of SWISS-PROT to have PTCs, rendering them potential targets of NMD. The precise correlation between LARD isoform expression and PTC+ status hints that there may be a role for alternative-splicing-induced NMD. Here, the gene structures of these five isoforms are shown alongside that of the full-length LARD isoform (isoform 1). In each case, the location of the stop codon has been labeled and, where appropriate, isoforms have been denoted as PTC+.
Similar articles
- UPF3 suppresses aberrant spliced mRNA in Arabidopsis.
Hori K, Watanabe Y. Hori K, et al. Plant J. 2005 Aug;43(4):530-40. doi: 10.1111/j.1365-313X.2005.02473.x. Plant J. 2005. PMID: 16098107 - Nanopore sequencing reveals endogenous NMD-targeted isoforms in human cells.
Karousis ED, Gypas F, Zavolan M, Mühlemann O. Karousis ED, et al. Genome Biol. 2021 Aug 13;22(1):223. doi: 10.1186/s13059-021-02439-3. Genome Biol. 2021. PMID: 34389041 Free PMC article. - [Progress on cis-acting regulatory elements in nonsense-mediated mRNA decay].
Huang Z, Zhou TH, Guo BJ. Huang Z, et al. Yi Chuan Xue Bao. 2004 Nov;31(11):1321-6. Yi Chuan Xue Bao. 2004. PMID: 15651687 Review. Chinese. - Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans.
Barberan-Soler S, Lambert NJ, Zahler AM. Barberan-Soler S, et al. RNA. 2009 Sep;15(9):1652-60. doi: 10.1261/rna.1711109. Epub 2009 Jul 17. RNA. 2009. PMID: 19617316 Free PMC article. - The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision?
Silva AL, Romão L. Silva AL, et al. FEBS Lett. 2009 Feb 4;583(3):499-505. doi: 10.1016/j.febslet.2008.12.058. Epub 2009 Jan 20. FEBS Lett. 2009. PMID: 19162024 Review.
Cited by
- Gene expression: sizing it all up.
Woody JL, Shoemaker RC. Woody JL, et al. Front Genet. 2011 Oct 17;2:70. doi: 10.3389/fgene.2011.00070. eCollection 2011. Front Genet. 2011. PMID: 22303365 Free PMC article. - Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing.
Xing Y, Lee CJ. Xing Y, et al. PLoS Genet. 2005 Sep;1(3):e34. doi: 10.1371/journal.pgen.0010034. PLoS Genet. 2005. PMID: 16170410 Free PMC article. - Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner.
Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Avery P, et al. RNA. 2011 Apr;17(4):624-38. doi: 10.1261/rna.2404211. Epub 2011 Feb 11. RNA. 2011. PMID: 21317294 Free PMC article. - The contribution of Alu exons to the human proteome.
Lin L, Jiang P, Park JW, Wang J, Lu ZX, Lam MP, Ping P, Xing Y. Lin L, et al. Genome Biol. 2016 Jan 28;17:15. doi: 10.1186/s13059-016-0876-5. Genome Biol. 2016. PMID: 26821878 Free PMC article. - A genome-wide survey demonstrates widespread non-linear mRNA in expressed sequences from multiple species.
Dixon RJ, Eperon IC, Hall L, Samani NJ. Dixon RJ, et al. Nucleic Acids Res. 2005 Oct 19;33(18):5904-13. doi: 10.1093/nar/gki893. Print 2005. Nucleic Acids Res. 2005. PMID: 16237125 Free PMC article.
References
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K22 HG000056/HG/NHGRI NIH HHS/United States
- T32 HG000047/HG/NHGRI NIH HHS/United States
- K22 HG00056/HG/NHGRI NIH HHS/United States
- T32 HG00047/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials