High-resolution analysis of DNA copy number using oligonucleotide microarrays - PubMed (original) (raw)
Comparative Study
doi: 10.1101/gr.2012304.
Jing Huang, Joel Greshock, Stephen Watt, Adam Butler, Sofie West, Mira Grigorova, Keith W Jones, Wen Wei, Michael R Stratton, P Andrew Futreal, Barbara Weber, Michael H Shapero, Richard Wooster
Affiliations
- PMID: 14762065
- PMCID: PMC327104
- DOI: 10.1101/gr.2012304
Comparative Study
High-resolution analysis of DNA copy number using oligonucleotide microarrays
Graham R Bignell et al. Genome Res. 2004 Feb.
Abstract
Genomic copy number alterations are a feature of many human diseases including cancer. We have evaluated the effectiveness of an oligonucleotide array, originally designed to detect single-nucleotide polymorphisms, to assess DNA copy number. We first showed that fluorescent signal from the oligonucleotide array varies in proportion to both decreases and increases in copy number. Subsequently we applied the system to a series of 20 cancer cell lines. All of the putative homozygous deletions (10) and high-level amplifications (12; putative copy number >4) tested were confirmed by PCR (either qPCR or normal PCR) analysis. Low-level copy number changes for two of the lines under analysis were compared with BAC array CGH; 77% (n = 44) of the autosomal chromosomes used in the comparison showed consistent patterns of LOH (loss of heterozygosity) and low-level amplification. Of the remaining 10 comparisons that were discordant, eight were caused by low SNP densities and failed in both lines. The studies demonstrate that combining the genotype and copy number analyses gives greater insight into the underlying genetic alterations in cancer cells with identification of complex events including loss and reduplication of loci.
Figures
Figure 1
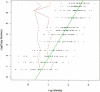
Plot of log(copy number) against log(intensity) for the spiking experiment in which 18 aliquots of the same DNA were spiked with varying concentrations of 42 SNPs from twofold up to 1000-fold. The black dots indicate the results for the individual SNPs across the 12 spiking concentrations (spiked with an extra 0, 2, 5, 10, 25, 50, 75, 100, 250, 500, 750, and 1000 copies); the green dots and line show the mean for all 42 SNPs. Two SNPs did not report increased fluorescence with increased copy number, and these are highlighted in red.
Figure 2
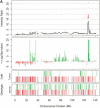
Examples of amplification and deletions together with genotyping data generated by the p501 array compared with BAC CGH data. (Top panel) The fluorescence ratio plot from the p501 array showing the smoothed fluorescent intensity data of the sample divided by the figure from the reference sample. (Second panel) The _p_-value plot for the individual SNPs calculated by comparison to the mean and standard deviation of 29 normal DNAs with deletions represented by red lines and amplifications in green. (Third and fourth panels) The genotypes for the tumor and matched normal samples, respectively; homozygous SNPs are represented by a red line below the center point whereas heterozygous markers are represented by a green line above the center point. (Bottom panel in C and D only) The results for BAC-array-based CGH (where available). (A) Genomic amplification at the C-MYC locus (arrow) on Chromosome 8 in the prostate line COR-L96-CAR. (B) A homozygous deletion (arrow) at the p16/INK4 locus on Chromosome 9 in the renal cell carcinoma cell line LB1047. (C) Chromosome 18 from the breast cancer cell line HCC1937. (i) This chromosome has a copy number of 2 (intensity ratio of 1) from the pter until 18q12.1. (ii) The copy number drops to 1 (intensity ratio of 0.5) from 18q12.1 until the 18qter, although the genotyping data indicate that this line is heterozygous across the full length of Chromosome 18. (D) Chromosome 5 from the small cell lung cancer line NCI-H209. This chromosome shows a complex pattern with partial amplification of the p-arm (i), followed by a drop in fluorescent intensity to 0.5 with corresponding LOH determined from the genotyping data (ii), until 5q23.2 (iii), where the intensity ratio recovers to 1 (copy number of 2); however, this region still represents LOH as determined by the genotyping data and therefore represents duplication of a single parental chromosome. There is also a homozygous deletion at 5q14.3 (arrow). This was not detected with the BAC array, as the array does not have a clone that covers this region.
Figure 2
Examples of amplification and deletions together with genotyping data generated by the p501 array compared with BAC CGH data. (Top panel) The fluorescence ratio plot from the p501 array showing the smoothed fluorescent intensity data of the sample divided by the figure from the reference sample. (Second panel) The _p_-value plot for the individual SNPs calculated by comparison to the mean and standard deviation of 29 normal DNAs with deletions represented by red lines and amplifications in green. (Third and fourth panels) The genotypes for the tumor and matched normal samples, respectively; homozygous SNPs are represented by a red line below the center point whereas heterozygous markers are represented by a green line above the center point. (Bottom panel in C and D only) The results for BAC-array-based CGH (where available). (A) Genomic amplification at the C-MYC locus (arrow) on Chromosome 8 in the prostate line COR-L96-CAR. (B) A homozygous deletion (arrow) at the p16/INK4 locus on Chromosome 9 in the renal cell carcinoma cell line LB1047. (C) Chromosome 18 from the breast cancer cell line HCC1937. (i) This chromosome has a copy number of 2 (intensity ratio of 1) from the pter until 18q12.1. (ii) The copy number drops to 1 (intensity ratio of 0.5) from 18q12.1 until the 18qter, although the genotyping data indicate that this line is heterozygous across the full length of Chromosome 18. (D) Chromosome 5 from the small cell lung cancer line NCI-H209. This chromosome shows a complex pattern with partial amplification of the p-arm (i), followed by a drop in fluorescent intensity to 0.5 with corresponding LOH determined from the genotyping data (ii), until 5q23.2 (iii), where the intensity ratio recovers to 1 (copy number of 2); however, this region still represents LOH as determined by the genotyping data and therefore represents duplication of a single parental chromosome. There is also a homozygous deletion at 5q14.3 (arrow). This was not detected with the BAC array, as the array does not have a clone that covers this region.
Figure 2
Examples of amplification and deletions together with genotyping data generated by the p501 array compared with BAC CGH data. (Top panel) The fluorescence ratio plot from the p501 array showing the smoothed fluorescent intensity data of the sample divided by the figure from the reference sample. (Second panel) The _p_-value plot for the individual SNPs calculated by comparison to the mean and standard deviation of 29 normal DNAs with deletions represented by red lines and amplifications in green. (Third and fourth panels) The genotypes for the tumor and matched normal samples, respectively; homozygous SNPs are represented by a red line below the center point whereas heterozygous markers are represented by a green line above the center point. (Bottom panel in C and D only) The results for BAC-array-based CGH (where available). (A) Genomic amplification at the C-MYC locus (arrow) on Chromosome 8 in the prostate line COR-L96-CAR. (B) A homozygous deletion (arrow) at the p16/INK4 locus on Chromosome 9 in the renal cell carcinoma cell line LB1047. (C) Chromosome 18 from the breast cancer cell line HCC1937. (i) This chromosome has a copy number of 2 (intensity ratio of 1) from the pter until 18q12.1. (ii) The copy number drops to 1 (intensity ratio of 0.5) from 18q12.1 until the 18qter, although the genotyping data indicate that this line is heterozygous across the full length of Chromosome 18. (D) Chromosome 5 from the small cell lung cancer line NCI-H209. This chromosome shows a complex pattern with partial amplification of the p-arm (i), followed by a drop in fluorescent intensity to 0.5 with corresponding LOH determined from the genotyping data (ii), until 5q23.2 (iii), where the intensity ratio recovers to 1 (copy number of 2); however, this region still represents LOH as determined by the genotyping data and therefore represents duplication of a single parental chromosome. There is also a homozygous deletion at 5q14.3 (arrow). This was not detected with the BAC array, as the array does not have a clone that covers this region.
Figure 2
Examples of amplification and deletions together with genotyping data generated by the p501 array compared with BAC CGH data. (Top panel) The fluorescence ratio plot from the p501 array showing the smoothed fluorescent intensity data of the sample divided by the figure from the reference sample. (Second panel) The _p_-value plot for the individual SNPs calculated by comparison to the mean and standard deviation of 29 normal DNAs with deletions represented by red lines and amplifications in green. (Third and fourth panels) The genotypes for the tumor and matched normal samples, respectively; homozygous SNPs are represented by a red line below the center point whereas heterozygous markers are represented by a green line above the center point. (Bottom panel in C and D only) The results for BAC-array-based CGH (where available). (A) Genomic amplification at the C-MYC locus (arrow) on Chromosome 8 in the prostate line COR-L96-CAR. (B) A homozygous deletion (arrow) at the p16/INK4 locus on Chromosome 9 in the renal cell carcinoma cell line LB1047. (C) Chromosome 18 from the breast cancer cell line HCC1937. (i) This chromosome has a copy number of 2 (intensity ratio of 1) from the pter until 18q12.1. (ii) The copy number drops to 1 (intensity ratio of 0.5) from 18q12.1 until the 18qter, although the genotyping data indicate that this line is heterozygous across the full length of Chromosome 18. (D) Chromosome 5 from the small cell lung cancer line NCI-H209. This chromosome shows a complex pattern with partial amplification of the p-arm (i), followed by a drop in fluorescent intensity to 0.5 with corresponding LOH determined from the genotyping data (ii), until 5q23.2 (iii), where the intensity ratio recovers to 1 (copy number of 2); however, this region still represents LOH as determined by the genotyping data and therefore represents duplication of a single parental chromosome. There is also a homozygous deletion at 5q14.3 (arrow). This was not detected with the BAC array, as the array does not have a clone that covers this region.
Figure 3
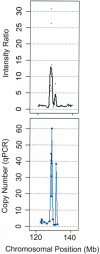
Comparison of copy number estimation for the amplification of the c-MYC proto-oncogene in COR-L96-CAR using the p501 array (A; where copy number equals two times the intensity ratio) and qPCR (B).
Similar articles
- Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.
Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Kloth JN, et al. BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article. - Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex changes and multiple forms of chromosomal instability in colorectal cancers.
Gaasenbeek M, Howarth K, Rowan AJ, Gorman PA, Jones A, Chaplin T, Liu Y, Bicknell D, Davison EJ, Fiegler H, Carter NP, Roylance RR, Tomlinson IP. Gaasenbeek M, et al. Cancer Res. 2006 Apr 1;66(7):3471-9. doi: 10.1158/0008-5472.CAN-05-3285. Cancer Res. 2006. PMID: 16585170 - Single nucleotide polymorphism microarray analysis of genetic alterations in cancer.
Mullighan CG. Mullighan CG. Methods Mol Biol. 2011;730:235-58. doi: 10.1007/978-1-61779-074-4_17. Methods Mol Biol. 2011. PMID: 21431646 - SNP array analysis in hematologic malignancies: avoiding false discoveries.
Heinrichs S, Li C, Look AT. Heinrichs S, et al. Blood. 2010 May 27;115(21):4157-61. doi: 10.1182/blood-2009-11-203182. Epub 2010 Mar 19. Blood. 2010. PMID: 20304806 Free PMC article. Review. - The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer.
Lapunzina P, Monk D. Lapunzina P, et al. Biol Cell. 2011 Jul;103(7):303-17. doi: 10.1042/BC20110013. Biol Cell. 2011. PMID: 21651501 Review.
Cited by
- Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue.
Mayrhofer M, DiLorenzo S, Isaksson A. Mayrhofer M, et al. Genome Biol. 2013 Mar 25;14(3):R24. doi: 10.1186/gb-2013-14-3-r24. Genome Biol. 2013. PMID: 23531354 Free PMC article. - arrayMap: a reference resource for genomic copy number imbalances in human malignancies.
Cai H, Kumar N, Baudis M. Cai H, et al. PLoS One. 2012;7(5):e36944. doi: 10.1371/journal.pone.0036944. Epub 2012 May 18. PLoS One. 2012. PMID: 22629346 Free PMC article. - Advances in understanding cancer genomes through second-generation sequencing.
Meyerson M, Gabriel S, Getz G. Meyerson M, et al. Nat Rev Genet. 2010 Oct;11(10):685-96. doi: 10.1038/nrg2841. Nat Rev Genet. 2010. PMID: 20847746 Review. - The impact of genomic alterations on the transcriptome: a prostate cancer cell line case study.
Chaudhary J, Schmidt M. Chaudhary J, et al. Chromosome Res. 2006;14(5):567-86. doi: 10.1007/s10577-006-1055-4. Epub 2006 Jul 12. Chromosome Res. 2006. PMID: 16823619
References
- Albertson, D.G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S.H., Kowbel, D., Kuo, W.L., Gray, J.W., and Pinkel, D. 2000. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 25: 144-146. - PubMed
- Dumur, C.I., Dechsukhum, C., Ware, J.L., Cofield, S.S., Best, A.M., Wilkinson, D.S., Garrett, C.T., and Ferreira-Gonzalez, A. 2003. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics 81: 260-269. - PubMed
- Hodgson, G., Hager, J.H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D.G., Pinkel, D., Collins, C., et al. 2001. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 29: 459-464. - PubMed
- Jain, A.N., Chin, K., Borresen-Dale, A.L., Erikstein, B.K., Eynstein Lonning, P., Kaaresen, R., and Gray, J.W. 2001. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc. Natl. Acad. Sci. 98: 7952-7957. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials