Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin - PubMed (original) (raw)
Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin
Abarna Thiru et al. EMBO J. 2004.
Abstract
HP1 family proteins are adaptor molecules, containing two related chromo domains that are required for chromatin packaging and gene silencing. Here we present the structure of the chromo shadow domain from mouse HP1beta bound to a peptide containing a consensus PXVXL motif found in many HP1 binding partners. The shadow domain exhibits a novel mode of peptide recognition, where the peptide binds across the dimer interface, sandwiched in a beta-sheet between strands from each monomer. The structure allows us to predict which other shadow domains bind similar PXVXL motif-containing peptides and provides a framework for predicting the sequence specificity of the others. We show that targeting of HP1beta to heterochromatin requires shadow domain interactions with PXVXL-containing proteins in addition to chromo domain recognition of Lys-9-methylated histone H3. Interestingly, it also appears to require the simultaneous recognition of two Lys-9-methylated histone H3 molecules. This finding implies a further complexity to the histone code for regulation of chromatin structure and suggests how binding of HP1 family proteins may lead to its condensation.
Figures
Figure 1
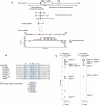
The HP1 interaction region, located within the N-terminal domain of CAF-1 p150, binds to the shadow domain of mouse HP1β. (A) A number of overlapping cDNA clones encoding portions of mouse CAF-1 p150 were isolated through a two-hybrid screen using HP1β as bait (Murzina et al, 1999). These clones had a 66-residue sequence in common that was expressed and used in limited proteolysis experiments with HP1C to define the interacting fragment. This was subsequently refined using NMR chemical shift mapping and 15N NMR relaxation experiments. The residues coloured red are involved in hydrophobic interactions with the HP1β shadow domain. (B) Sequence alignment of (i) proteins that contain the PXVXL motif (conserved residues coloured red) and (ii) peptides identified in phage display experiments (Smothers and Henikoff, 2000). The positions of residues involved in the interaction with CAF-1 are highlighted in blue. (C) Example regions of the 13C, 15N X-filtered NOESY spectrum illustrating the different intermolecular NOEs observed to the CAF-1 peptide from L168 Hδ1 in the two HP1C monomers (A, B).
Figure 2
Structure of the HP1β shadow domain/CAF-1 complex. (A) The backbone of residues 110–172 of HP1β and 214–232 of CAF-1 from the 25 lowest energy structures (out of 72 that converged from the 100 computed). The structure has good covalent geometry and nonbonded contacts (Table 1). The peptide backbone around residues Asp-214–Glu-219 of CAF-1 is not well defined in the structure because only the Phe-217 and Ile-218 side chains interact with the shadow domain. 15N relaxation studies show that the peptide backbone in this region has increased mobility within the complex when compared to residues in the PXVXL motif (Figure 1A). (B) The structure closest to the mean, rotated by 90° about the _z_-axis, compared to the orientation shown in (A). This structure is compared with that of the free shadow domain (PDB code: 1DZ1; Brasher et al, 2000) and the chromo domain/histone H3 complex (PDB code: 1GUW; Nielsen et al, 2002). In the structure of the free shadow domain, the side chains of Trp-170, which stabilise the position of the C-terminal tails, are shown. (C) Stereoview of the structure closest to the mean showing side chains of residues involved in the HP1β/CAF-1 peptide interface. (D, E) Close-up views of the interactions made by (D) Val-224 (position 0) and (E) Pro-222 and Leu-226 (positions −2/+2), Phe-217/Ile-218 (−7/−6 positions) and Ile-229/Leu-230 (+5/+6 positions). In (A–D), the HP1β monomers are coloured blue and magenta and the CAF-1 peptide is coloured green. (Note: Trp-170 is not shown on monomer A because it does not interact with Leu-226 at the +2 position.)
Figure 3
Shadow domain alignment in which the residues involved in complex formation are highlighted (numbering is for mouse HP1β). Residues important for recognition of the PXVXL motif are highlighted in yellow (Val-224) and red (Pro-222/Leu-226 at the −2/+2 positions). Residues that form the hydrophobic patch that interacts with the flanking N- and C-terminal sequences (Phe-217/Ile-218 and Ile-229/Leu-230 at the −6/−7 and +5/+6 positions, respectively) are highlighted in blue. Residues that are not conserved and are predicted to alter specificity are boxed. Conserved residues that define the fold of the shadow domain are highlighted in grey, while residues important for dimerisation are indicated by blue triangles. Phe-163 (indicated by a green dot) is important for both the structure of the shadow domain and peptide binding. The positions of secondary structure elements in mouse HP1β are indicated by purple arrows (β-strands) and cylinders (α-helices)—the dots indicate the positions of the conserved bulges in the first strand. Sequences are labelled by species name (Mm—Mus musculus, Hs—Homo sapiens, Gg—Gallus gallus, Xl—Xenopus laevis, Dm—Drosophila melanogaster, Dv—Drosophila virilis, Os—Oryza sativa, Zm—Zea mays, At—Arabidopsis thaliana, Dc—Daucus carota, Sp—Schizosaccharomyces pombe, Ec—Encephalitozoon cuniculi, Ce—Caenorhabditis elegans, Le—Lycopersicon esculentum).
Figure 4
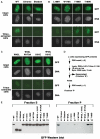
Localisation of HP1β to heterochromatin. Mouse L cells were transfected with constructs expressing full-length GFP-HP1β, GFP-chromo domain (residues 10–80) or GFP-shadow domain (residues 104–185). Heterochromatin targeting was assessed by staining DNA with limiting amounts of Hoechst 33258. (A) Localisation of GFP full-length HP1β compared with that of either the chromo or shadow domain alone. (B) Localisation of GFP full-length HP1β with point mutations that impair binding to all PXVXL proteins (W170A and L168 H) or affect the charge of Thr-169 and thereby weaken binding to some PXVXL proteins (T169D and T169 K). (C) Localisation of GFP full-length HP1β with point mutations that cripple binding to Lys-9-methylated histone H3 (W42L), Lys-9-methylated histone H3 and PXVXL proteins (W42L, W170), dimerisation and binding to PXVXL proteins (I161E), and dimerisation and binding to PXVXL proteins and binding to Lys-9-methylated histone H3 (W42L, I161E). In (A–C), a comparison is shown between intact cells and cells that have been permeabilised with Triton and washed. Exposure was 10 times longer for the Triton-extracted cells. (D) Experimental scheme of the Western blot in (E). (E) Western blot analysis (using an anti-GFP antibody) of GFP fusion proteins in the supernatant (S) and pellet (P) fractions following Triton permeabilisation and extraction of transfected cells.
Similar articles
- The HP1 chromo shadow domain binds a consensus peptide pentamer.
Smothers JF, Henikoff S. Smothers JF, et al. Curr Biol. 2000 Jan 13;10(1):27-30. doi: 10.1016/s0960-9822(99)00260-2. Curr Biol. 2000. PMID: 10660299 - The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer.
Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. Brasher SV, et al. EMBO J. 2000 Apr 3;19(7):1587-97. doi: 10.1093/emboj/19.7.1587. EMBO J. 2000. PMID: 10747027 Free PMC article. - Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
Liu Y, Qin S, Lei M, Tempel W, Zhang Y, Loppnau P, Li Y, Min J. Liu Y, et al. J Biol Chem. 2017 Apr 7;292(14):5655-5664. doi: 10.1074/jbc.M116.768374. Epub 2017 Feb 21. J Biol Chem. 2017. PMID: 28223359 Free PMC article. - Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly.
Nishibuchi G, Nakayama J. Nishibuchi G, et al. J Biochem. 2014 Jul;156(1):11-20. doi: 10.1093/jb/mvu032. Epub 2014 May 13. J Biochem. 2014. PMID: 24825911 Review. - How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.
Sales-Gil R, Vagnarelli P. Sales-Gil R, et al. Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review.
Cited by
- Heterochromatin protein 1 alpha (HP1α) undergoes a monomer to dimer transition that opens and compacts live cell genome architecture.
Lou J, Deng Q, Zhang X, Bell CC, Das AB, Bediaga NG, Zlatic CO, Johanson TM, Allan RS, Griffin MDW, Paradkar P, Harvey KF, Dawson MA, Hinde E. Lou J, et al. Nucleic Acids Res. 2024 Oct 14;52(18):10918-10933. doi: 10.1093/nar/gkae720. Nucleic Acids Res. 2024. PMID: 39193905 Free PMC article. - The tripartite motif-containing 24 is a multifunctional player in human cancer.
Yao Y, Zhou S, Yan Y, Fu K, Xiao S. Yao Y, et al. Cell Biosci. 2024 Aug 19;14(1):103. doi: 10.1186/s13578-024-01289-3. Cell Biosci. 2024. PMID: 39160596 Free PMC article. Review. - Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Shao Z, Lu J, Khudaverdyan N, Song J. Shao Z, et al. Nat Commun. 2024 Aug 9;15(1):6815. doi: 10.1038/s41467-024-51246-4. Nat Commun. 2024. PMID: 39122718 Free PMC article. - Epigenetic memory is governed by an effector recruitment specificity toggle in Heterochromatin Protein 1.
Ames A, Seman M, Larkin A, Raiymbek G, Chen Z, Levashkevich A, Kim B, Biteen JS, Ragunathan K. Ames A, et al. Nat Commun. 2024 Jul 25;15(1):6276. doi: 10.1038/s41467-024-50538-z. Nat Commun. 2024. PMID: 39054315 Free PMC article. - ChAHP2 and ChAHP control diverse retrotransposons by complementary activities.
Ahel J, Pandey A, Schwaiger M, Mohn F, Basters A, Kempf G, Andriollo A, Kaaij L, Hess D, Bühler M. Ahel J, et al. Genes Dev. 2024 Jul 19;38(11-12):554-568. doi: 10.1101/gad.351769.124. Genes Dev. 2024. PMID: 38960717 Free PMC article.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 3: 207–217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases