DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding - PubMed (original) (raw)
DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding
Dirk Remus et al. EMBO J. 2004.
Abstract
Drosophila origin recognition complex (ORC) localizes to defined positions on chromosomes, and in follicle cells the chorion gene amplification loci are well-studied examples. However, the mechanism of specific localization is not known. We have studied the DNA binding of DmORC to investigate the cis-requirements for DmORC:DNA interaction. DmORC displays at best six-fold differences in the relative affinities to DNA from the third chorion locus and to random fragments in vitro, and chemical probing and DNase1 protection experiments did not identify a discrete binding site for DmORC on any of these fragments. The intrinsic DNA-binding specificity of DmORC is therefore insufficient to target DmORC to origins of replication in vivo. However, the topological state of the DNA significantly influences the affinity of DmORC to DNA. We found that the affinity of DmORC for negatively supercoiled DNA is about 30-fold higher than for either relaxed or linear DNA. These data provide biochemical evidence for the notion that origin specification in metazoa likely involves mechanisms other than simple replicator-initiator interactions and that in vivo other proteins must determine ORC's localization.
Figures
Figure 1
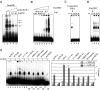
rDmORC binds to various DNA sequences. (A) EMSA showing the ATP-dependent binding of rDmORC to a radiolabeled ACE-3 fragment at increasing poly(dGdC)·poly(dGdC) competitor concentration as indicated. Lane 1, no protein. (B) EMSA showing the ATP-dependent binding of increasing amounts of rDmORC to ACE-3 at 50 μg/ml poly(dGdC)·poly(dGdC) competitor. (C) rDmORC–ACE-3 complex at equilibrium (lane 1) was chased with excess cold competitor DNA. Aliquots were analyzed by EMSA in intervals as indicated (lanes 2–4). *(s)=seconds (D) rDmORC binding to a radiolabeled ACE-3 fragment was monitored over time as indicated (lanes 2–4). Lane 1, no DmORC. (E) EMSA of rDmORC binding to various radiolabeled DNA fragments with or without 0.5 mM ATPγS as indicated. The bar diagram depicts binding efficiencies as a fraction of the bound probe. The averages and standard deviations for three independent experiments are shown.
Figure 2
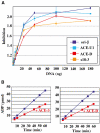
rDmORC binds with similar affinity to fragments of the third chromosome chorion gene cluster. ATP-dependent binding of rDmORC to a radiolabeled ACE-3 fragment was assayed by EMSA in the presence of competitor fragment DNA indicated in the upper right corner of each plot. % binding is relative to the fraction of ACE-3 bound in the absence of competitor. (A) Relative binding efficiencies of rDmORC (left two panels) and embryonic (0–12 h) DmORC to ACE-3 in the presence of increasing amounts of ACE-3 and ACE-D competitor. Dashed lines indicate [C]1/2. (B) Binding plots of all fragments tested. The relative position of each fragment in the chorion gene cluster is schematized in the center. (C) Summary plot depicting [C]1/2 for each fragment normalized to ACE-3. The average and standard deviation for two to three independent experiments are shown.
Figure 3
Inhibition of rDmORC's ATPase activity by DNA does not require a specific DNA sequence. (A) ATP hydrolysis by DmORC in the presence of increasing amounts of ori-β (dark blue line), ACE-U1 (light blue), ACE-D (red), and s18-3 (orange). The factor of inhibition is plotted as a function of fragment concentration. Averages and standard deviations for two independent experiments are shown. (B) Time course of the amount of ADP produced by rDmORC in the absence (blue lines) or presence (red lines) of ACE-3- (left panel) or ACE-D- (right panel) containing DNA.
Figure 4
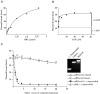
DmORC binds preferentially to (−) supercoiled DNA. rDmORC binding to 125 fmol 3H-labeled pBS/ori-β was assayed by nitrocellulose filter binding. (A) Titration of rDmORC with 0.5 mM ATP. (B) Titration of ATP with 1 pmol rDmORC. (C) rDmORC binding to 3H-labeled (−) supercoiled pBS/ori-β in the presence of increasing amounts of unlabeled (−) supercoiled pBS/ori-β (▪), pBS/ori-β relaxed with _Topo_I (○), pBS/ori-β linearized with _Bam_HI (×), or (−) supercoiled pBluescript (▵). Inset panel: Ethidium-bromide-stained agarose gel showing (−) supercoiled, linearized, and relaxed pBS/ori-β.
Figure 5
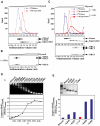
Superhelical density determines the preference for DmORC–DNA binding. Binding reactions containing equimolar 32P-labeled linear and 3H-labeled (−) supercoiled DNA were fractionated by glycerol gradient sedimentation and assayed for each radiolabel by liquid scintillation counting, analyzed by Southern blotting for pBS/ori-β, and by Western blotting for rDmORC subunits 1, 2, and 6. (A) Upper panel: sedimentation profile of linear (blue dashed line) and (−) supercoiled (red dashed line) DNA in the absence of rDmORC. Lower panel: Southern blot of the same fractions. (B) Western blot: sedimentation of rDmORC without DNA. (C) Sedimentation profile of linear (blue) and (−) supercoiled (red) DNA in the absence (dashed lines) and presence (solid lines) of rDmORC. Middle panel: Southern blot analysis of binding reaction; lower panel: Western blot, DmORC sedimentation in binding reaction. (D) rDmORC binding to (−) supercoiled DNA of variable σ. An ethidium-bromide-stained gel of the tested DNAs is shown on top of a graph depicting the ratios of bound supercoiled DNA to bound linear DNA as a function of σ of input supercoiled DNA. (E) Relative binding efficiency of (+) supercoiled DNA (red column) compared to (−) supercoiled DNA of bracketing superhelical densities. An ethidium-bromide-stained gel of relaxed, (−) supercoiled, and (+) supercoiled DNA is shown on top. M=1 kb ladder.
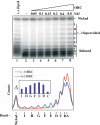
Figure 6
rDmORC binding to (−) supercoiled DNA induces a topology change in the DNA. A measure of 250 fmol of (−) supercoiled pBS/ori-β was incubated with increasing amounts of rDmORC in the presence of _Topo_I, the DNA repurified and analyzed by Southern blotting (upper panel). Lane 1, no _Topo_I; Lane 2, no rDmORC. Lower panel: lane scan of lanes −ORC (red) and +0.8 μg ORC (blue). Inset: Difference plot of topoisomer band intensities from Southern blot lanes 8 and 2. The letters denote topoisomers.
Similar articles
- Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element.
Austin RJ, Orr-Weaver TL, Bell SP. Austin RJ, et al. Genes Dev. 1999 Oct 15;13(20):2639-49. doi: 10.1101/gad.13.20.2639. Genes Dev. 1999. PMID: 10541550 Free PMC article. - CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex.
Remus D, Blanchette M, Rio DC, Botchan MR. Remus D, et al. J Biol Chem. 2005 Dec 2;280(48):39740-51. doi: 10.1074/jbc.M508515200. Epub 2005 Sep 27. J Biol Chem. 2005. PMID: 16188887 - Sequence requirements for function of the Drosophila chorion gene locus ACE3 replicator and ori-beta origin elements.
Zhang H, Tower J. Zhang H, et al. Development. 2004 May;131(9):2089-99. doi: 10.1242/dev.01064. Development. 2004. PMID: 15105371 - Developmental gene amplification and origin regulation.
Tower J. Tower J. Annu Rev Genet. 2004;38:273-304. doi: 10.1146/annurev.genet.37.110801.143851. Annu Rev Genet. 2004. PMID: 15568978 Review. - ORC binding, gene amplification, and the nature of metazoan replication origins.
Spradling AC. Spradling AC. Genes Dev. 1999 Oct 15;13(20):2619-23. doi: 10.1101/gad.13.20.2619. Genes Dev. 1999. PMID: 10541547 Review. No abstract available.
Cited by
- Controlled interconversion of macrocyclic atropisomers via defined intermediates.
Sun X, Bai JK, Yang YD, Zhu KL, Liang JQ, Wang XY, Xiang JF, Hao X, Liang TL, Guan AJ, Wu NN, Gong HY. Sun X, et al. Nat Commun. 2024 Aug 2;15(1):6559. doi: 10.1038/s41467-024-50739-6. Nat Commun. 2024. PMID: 39095340 Free PMC article. - Multiple pathways for licensing human replication origins.
Yang R, Hunker O, Wise M, Bleichert F. Yang R, et al. bioRxiv [Preprint]. 2024 Apr 10:2024.04.10.588796. doi: 10.1101/2024.04.10.588796. bioRxiv. 2024. PMID: 38645015 Free PMC article. Preprint. - Activity of zebrafish THAP9 transposase and zebrafish P element-like transposons.
Kutnowski N, Ghanim GE, Lee Y, Rio DC. Kutnowski N, et al. bioRxiv [Preprint]. 2024 Mar 22:2024.03.22.586318. doi: 10.1101/2024.03.22.586318. bioRxiv. 2024. PMID: 38562726 Free PMC article. Preprint. - The origin recognition complex requires chromatin tethering by a hypervariable intrinsically disordered region that is functionally conserved from sponge to man.
Adiji OA, McConnell BS, Parker MW. Adiji OA, et al. Nucleic Acids Res. 2024 May 8;52(8):4344-4360. doi: 10.1093/nar/gkae122. Nucleic Acids Res. 2024. PMID: 38381902 Free PMC article. - DNA mimic foldamers affect chromatin composition and disturb cell cycle progression.
Kleene V, Corvaglia V, Chacin E, Forne I, Konrad DB, Khosravani P, Douat C, Kurat CF, Huc I, Imhof A. Kleene V, et al. Nucleic Acids Res. 2023 Oct 13;51(18):9629-9642. doi: 10.1093/nar/gkad681. Nucleic Acids Res. 2023. PMID: 37650653 Free PMC article.
References
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 - PubMed
- Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16: 659–672 - PubMed
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases