New perspective on uncultured bacterial phylogenetic division OP11 - PubMed (original) (raw)
New perspective on uncultured bacterial phylogenetic division OP11
J Kirk Harris et al. Appl Environ Microbiol. 2004 Feb.
Abstract
Organisms belonging to the OP11 candidate phylogenetic division of Bacteria have been detected only in rRNA-based sequence surveys of environmental samples. Preliminary studies indicated that such organisms represented by the sequences are abundant and widespread in nature and highly diverse phylogenetically. In order to document more thoroughly the phylogenetic breadth and environmental distribution of this diverse group of organisms, we conducted further molecular analyses on environmental DNAs. Using PCR techniques and primers directed toward each of the five described subdivisions of OP11, we surveyed 17 environmental DNAs and analyzed rRNA gene sequences in 27 clonal libraries from 14 environments. Ninety-nine new and unique sequences were determined completely, and approximately 200 additional clones were subjected to partial sequencing. Extensive phylogenetic comparisons of the new sequences to those representing other bacterial divisions further resolved the phylogeny of the bacterial candidate division OP11 and identified two new candidate bacterial divisions, OP11-derived 1 (OD1) and Sulphur River 1 (SR1). The widespread environmental distribution of representatives of the bacterial divisions OD1, OP11, and SR1 suggests potentially conspicuous biogeochemical roles for these organisms in their respective environments. The information on environmental distribution offers clues for attempts to culture landmark representatives of these novel bacterial divisions, and the sequences are specific molecular signatures that provide for their identification in other contexts.
Figures
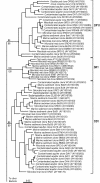
FIG. 1.
Bootstrap consensus tree showing the well-supported phylogenetic relationships for the bacterial divisions OP11, OD1, and SR1, with division names listed outside the brackets. Nodes within the tree that are supported (bootstrap values of >70%) (9) in all analyses (distance, MP, and ML [see Results]) are indicated by filled circles. Nodes without circles were not strongly supported (bootstrap of <70%) in all analyses. Bootstrap support for the division-level divergences (Table 2) are given for ME (top, 245-taxa data set, HKY85 model of evolution) and MP (bottom, 245-taxa data set). The bar represents 5% sequence divergence.
Similar articles
- Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria.
Dojka MA, Harris JK, Pace NR. Dojka MA, et al. Appl Environ Microbiol. 2000 Apr;66(4):1617-21. doi: 10.1128/AEM.66.4.1617-1621.2000. Appl Environ Microbiol. 2000. PMID: 10742250 Free PMC article. - Assessment of the diversity, abundance, and ecological distribution of members of candidate division SR1 reveals a high level of phylogenetic diversity but limited morphotypic diversity.
Davis JP, Youssef NH, Elshahed MS. Davis JP, et al. Appl Environ Microbiol. 2009 Jun;75(12):4139-48. doi: 10.1128/AEM.00137-09. Epub 2009 Apr 24. Appl Environ Microbiol. 2009. PMID: 19395567 Free PMC article. - Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation.
Dojka MA, Hugenholtz P, Haack SK, Pace NR. Dojka MA, et al. Appl Environ Microbiol. 1998 Oct;64(10):3869-77. doi: 10.1128/AEM.64.10.3869-3877.1998. Appl Environ Microbiol. 1998. PMID: 9758812 Free PMC article. - Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments.
Webster G, Parkes RJ, Fry JC, Weightman AJ. Webster G, et al. Appl Environ Microbiol. 2004 Sep;70(9):5708-13. doi: 10.1128/AEM.70.9.5708-5713.2004. Appl Environ Microbiol. 2004. PMID: 15345467 Free PMC article. - Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences.
Schmidt TM, Relman DA. Schmidt TM, et al. Methods Enzymol. 1994;235:205-22. doi: 10.1016/0076-6879(94)35142-2. Methods Enzymol. 1994. PMID: 7520119 Review. No abstract available.
Cited by
- Culture independent molecular analysis of bacterial communities in the mangrove sediment of Sundarban, India.
Ghosh A, Dey N, Bera A, Tiwari A, Sathyaniranjan KB, Chakrabarti K, Chattopadhyay D. Ghosh A, et al. Saline Syst. 2010 Feb 17;6(1):1. doi: 10.1186/1746-1448-6-1. Saline Syst. 2010. PMID: 20163727 Free PMC article. - Miniprimer PCR, a new lens for viewing the microbial world.
Isenbarger TA, Finney M, Ríos-Velázquez C, Handelsman J, Ruvkun G. Isenbarger TA, et al. Appl Environ Microbiol. 2008 Feb;74(3):840-9. doi: 10.1128/AEM.01933-07. Epub 2007 Dec 14. Appl Environ Microbiol. 2008. PMID: 18083877 Free PMC article. - A comparison of methods for total community DNA preservation and extraction from various thermal environments.
Mitchell KR, Takacs-Vesbach CD. Mitchell KR, et al. J Ind Microbiol Biotechnol. 2008 Oct;35(10):1139-47. doi: 10.1007/s10295-008-0393-y. Epub 2008 Jul 17. J Ind Microbiol Biotechnol. 2008. PMID: 18633656 - Utilization of low-molecular-weight organic compounds by the filterable fraction of a lotic microbiome.
Ghuneim LJ, Distaso MA, Chernikova TN, Bargiela R, Lunev EA, Korzhenkov AA, Toshchakov SV, Rojo D, Barbas C, Ferrer M, Golyshina OV, Golyshin PN, Jones DL. Ghuneim LJ, et al. FEMS Microbiol Ecol. 2021 Feb 5;97(2):fiaa244. doi: 10.1093/femsec/fiaa244. FEMS Microbiol Ecol. 2021. PMID: 33264383 Free PMC article. - Beyond genomics in Patescibacteria: A trove of unexplored biology packed into ultrasmall bacteria.
Srinivas P, Peterson SB, Gallagher LA, Wang Y, Mougous JD. Srinivas P, et al. Proc Natl Acad Sci U S A. 2024 Dec 17;121(51):e2419369121. doi: 10.1073/pnas.2419369121. Epub 2024 Dec 12. Proc Natl Acad Sci U S A. 2024. PMID: 39665754 Free PMC article.
References
- Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. - PMC - PubMed
- Dalevi, D., P. Hugenholtz, and L. Blackall. 2001. A multiple-outgroup approach to resolving division-level phylogenetic relationships using 16S rDNA data. Int. J. Syst. Evol. Microbiol. 51:385-391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases