Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes - PubMed (original) (raw)
Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes
Kui Shin Voo et al. J Exp Med. 2004.
Abstract
The Epstein-Barr virus (EBV)-encoded nuclear antigen 1 (EBNA1) is expressed in all EBV-associated tumors, making it an important target for immunotherapy. However, evidence for major histocompatibility complex (MHC) class I-restricted EBNA1 peptides endogenously presented by EBV-transformed B and tumor cells remains elusive. Here we describe for the first time the identification of an endogenously processed human histocompatibility leukocyte antigen (HLA)-B8-restricted EBNA1 peptide that is recognized by CD8+ T cells. T cell recognition could be inhibited by the treatment of target cells with proteasome inhibitors that block the MHC class I antigen processing pathway, but not by an inhibitor (chloroquine) of MHC class II antigen processing. We also demonstrate that new protein synthesis is required for the generation of the HLA-B8 epitope for T cell recognition, suggesting that defective ribosomal products (DRiPs) are the major source of T cell epitopes. Experiments with protease inhibitors indicate that some serine proteases may participate in the degradation of EBNA1 DRiPs before they are further processed by proteasomes. These findings not only provide the first evidence of the presentation of an MHC class I-restricted EBNA1 epitope to CD8+ T cells, but also offer new insight into the molecular mechanisms involved in the processing and presentation of EBNA1.
Figures
Figure 1.
Generation of EBNA1-specific T cells. (A) T cells were generated from HLA-B8–expressing PBMCs of donor M after in vitro stimulation with synthetic peptides from EBNA1. 1.5 × 105 cells per well of PBMCs were used to generate EBNA1-P518–530 peptide–specific T cells. Peptides other than those used for repeated stimulations served as negative controls. For T cell recognition assays, peptides were pulsed onto 1359mel target cells and cocultured with T cells overnight. Cytokine release assays were performed as previously described (reference 11). (B) T cell recognition (T cell line M3-W1 from donor M) of 1359mel cells pulsed with EBNA1-P518–530 peptide was specifically inhibited by antibody against MHC class I molecules. T cell recognition assays were performed at an E/T ratio of 1:1. All results are expressed as IFN-γ release in picogram/milliliter and are the averages of duplicate values. All antibodies were used at a final concentration of 20 μg/ml each.
Figure 2.
Characterization of EBNA1-specific T cells. (A) Recognition of LCL 111 by T cell clones derived from the M3-W1 T cell line. (B) FACS® analysis of M3-W1-B9 T cells for CD8 expression. T cells were stained with anti–CD4-PE or anti–CD8-FITC. Positive staining for CD8 T cells is shown as an open histogram and control antibody staining is represented as a shaded histogram. (C) Identification of minimal EBNA1 T cell epitope for MHC class I binding. Four different peptides were made and pulsed onto 1359mel cells at 10 μM concentration. After washing, the peptide-pulsed cells were cocultured with T cells overnight. IFN-γ release was determined from culture supernatants. (D) EBNA1-P518–526 peptide titration experiment for M3W1-B9 T cell recognition. EBNA1-P518–526 peptide at various concentrations were pulsed on 1359mel cells and used as target cells to stimulate T cells. A control peptide EBNA1-P 572–584 was also used at various concentrations. Experiments were repeated twice with similar results.
Figure 3.
Natural processing and presentation of EBNA1 to M3-W1-B9 CD8+ T cells. (A) Recognition of full-length EBNA1-transfected 1359mel cells by M3-W1-B9 T cells. 1359mel cells were transfected with 200 ng EBNA1-GFP or EBNA1-GAr-del-GFP plasmid DNAs using LipofectAMINE. IFN-γ release was determined as described in Fig. 1. (B) Recognition of 1359 fibroblasts transfected with EBNA1-GFP by M3-W1-B9 T cells. 1359 fibroblasts were transfected with EBNA1 plasmid DNA by electroporation. T cell assays were performed at an E/T ratio of 2:1. (C) T cell recognition of autologous PBMCs infected with retroviral/EBNA1-GFP.
Figure 4.
HLA-B8 molecule functions as a restriction element for M3-W1-B9 CD8+ T cells. (A) T cell recognition of peptide-pulsed HLA-B8–expressing cell lines. (B) Identification of HLA-B8 molecule as a restriction element for T cell recognition. HEK293 cells cotransfected with HLA-B8 plus full-length EBNA1-GFP or EBNA1-GAr-del-GFP cDNAs (with GAr domain deleted) were tested for recognition by M3-W1-B9 CD8+ T cells. Positive and negative signs indicate cotransfection of target cells in the presence or absence of HLA-B8 cDNA, respectively. (C) Natural processing and presentation of the native form of EBNA1 for T cell recognition. HEK293 cells cotransfected with full-length EBNA1 and HLA-B8 cDNAs were cocultured with M3-W1-B9 CD8+ T cells overnight. IFN-γ secretion from T cells was determined by ELISA. (D) Endogenous generation of HLA-B8–restricted EBNA1 peptide for T cell recognition. HEK293 cells transfected with HLA-B8 cDNA were mixed with HEK293 cells transfected with EBNA1-GFP cDNA at a 1:1 ratio. The mixed cells were then cocultured with M3-W1-B9 CD8+ T cells overnight. IFN-γ release from CD8+ T cells was measured from culture supernatants.
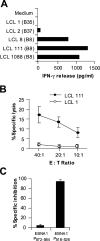
Figure 5.
Specific lysis of HLA-B8–matched EBV-transformed LCLs by M3-W1-B9 CD8+ T cells. (A) Recognition of HLA-B8–matched LCLs by M3-W1-B9 CD8+ T cells. LCLs were cocultured with M3-W1-B9 CD8+ T cells at an E/T ratio of 1:1. Mismatched LCLs were used as negative controls. (B) Specific lysis of HLA-B8–matched LCL 111 cells by CD8+ T cells at different E/T ratios. LCL 1 was used as a negative control. LCL cells were labeled with 51chromium. Cytolysis by CD8+ T cells was determined in a 16-h chromium release assay. (C) Cold target inhibition of recognition of LCL 111 cells by M3-W1-B9 CD8+ T cells. Lysis of LCLs by M3-W1-B9 CD8+ T cells was specifically inhibited when EBNA1-P518–526–pulsed cold LCL 111 targets were used. Lysis was tested with an effector to hot target ratio of 40:1. Cold LCL 111 target cells were pulsed with EBNA1-P518–526 or EBNA1-P572–584 peptide at a concentration of 1 μM and were mixed with hot targets at a ratio of 4:1. All experiments were repeated twice with similar results.
Figure 6.
Recognition of the EBNA1-P518–530 peptide by CD4+ and CD8+ T cells. (A) Alignment of HLA-DR1–, HLA-DP3–, and HLA-B8–restricted peptides. (B) Recognition of peptide-pulsed target cells by three different HLA-B8–, HLA-DR1–, and HLA-DP3–restricted T cell lines/clones. The HLA-DP3–restricted P3-B7 CD4+ T cells are described in a previous study (reference 11). The HLA-DR1–restricted A4.E116 CD4+ T cells, also previously described (reference 10), recognized an HLA-DR1–restricted EBNA1 peptide.
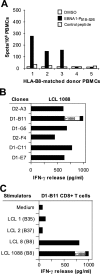
Figure 7.
Generation of EBNA1-P518–526 peptide–specific T cells from HLA-B8–expressing PBMCs. (A) Detection of EBNA1-P518–526 peptide–reactive T cells from HLA-B8+ donor PBMCs. 105 PBMCs were seeded per well and experiments were performed in quadruplicate wells. An HLA-A2–restricted NY-ESO-1 peptide served as a control. HLA-mismatched donor 5 and an HLA-B8+ PMBC donor 4 that is seronegative for EBV were also included. (B) Recognition of LCL 1088 by CD8+ T cell clones from the PBMCs of donor 3. T cells generated from PBMCs were stimulated with EBNA1-P518–526 peptide as described in Fig. 1. Six CD8+ T cell clones were generated from two T cell lines and were capable of recognizing HLA-B8–expressing LCL 1088 target cells. (C) D1-B11 CD8+ T cell recognition of HLA-B8–matched LCLs. LCLs were cocultured with T cells at an E/T ratio of 1:1. All experiments were repeated twice with similar results.
Figure 8.
Specific inhibition of T cell recognition of EBNA1 by proteasomes inhibitors. (A) Blocking of T cell recognition of EBNA1 by a ZAL proteasome inhibitor. HEK 293 cells cotransfected with EBNA1-GFP were treated with various concentrations of ZAL inhibitor for 10 h. After washing, cells were incubated with T cells overnight for IFN-γ release assays. Various dilutions of DMSO were used as controls. T cell activity in the absence of inhibitor was used at 100% activity. Two CD4+ T cells were used to demonstrate the specificity of ZAL inhibitor. TIL102 and P3-B7 CD4+ T cells able to recognize 102mel and HEK293/DP3/Ii-EBNA1 target cells, respectively, were not inhibited by ZAL. (B) Inhibition of M3-W1-B9 CD8+ T cell recognition of 1359mel target cells stably expressing EBNA1-GFP by lactacystin proteasome inhibitor. The lactacystin inhibitor did not affect recognition of 102mel tumor cells by TIL102 CD4+ cells. (C) Blocking of MHC class II antigen processing by chloroquine. Inhibition of T cell recognition of 102mel cells by TIL102 CD4+ was observed after treatment with chloroquine in a dose-dependent fashion. By contrast, T cell recognition of LCL 111 and HEK293 transfected with HLA-B8 and EBNA1-GFP cDNAs was not significantly affected after the treatment of chloroquine.
Figure 9.
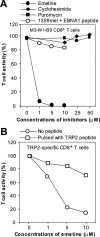
Inhibition of T cell recognition of EBNA1 by protein synthesis inhibitors. (A) Specific inhibition of M3-W1-B9 CD8+ T cell recognition of HEK293/B8/EBNA1-GFP target cells by an irreversible protein synthesis inhibitor emetine. HLA-B8–expressing HEK293/EBNA1-GFP target cells were treated with an emetine inhibitor at three concentrations for 1 h. After three washes, cells were cocultured with M3-W1-B9 CD8+ T cells overnight for IFN-γ release assays. Similar experiments were performed for the treatment of cells with cycloheximide or puromycin. To determine effect of emetine on recognition of MHC class I/EBNA1 peptide on the cell surface, we also pulsed HLA-B8+ 1359 cells with the EBNA1-P 518–526 peptide after the treatment of 1359mel cells with three different concentration of emetine. (B) Determination of the sensitivity of recognition of TRP2-specific CD8+ T cells to the treatment with emetine. 1363mel cells were treated with three different concentrations of emetine. After three washes, the cells were cocultured with TRP2-specific CD8+ T cells. The treated cells pulsed with a TRP2 peptide were used to examine the effect of emetine on recognition of MHC class I–TRP2 complexes on the cell surface.
Figure 10.
Requirement of serine proteases for the generation of EBNA1 T cell epitope. (A) Specific inhibition of M3-W1-B9 CD8+ T cell recognition of HEK293/B8/EBNA1-GFP target cells by protease inhibitors. Target cells were incubated with various protease inhibitors for 2 h, washed, and cocultured with CD8+ T cells overnight for IFN-γ release assays. Solvents used to solubilize inhibitors were also used as controls. Experiments were performed in triplicate wells. CD8+ T cell recognition of target cells was inhibited by treatment with TPCK and AEBSF inhibitors. (B) Effect of protease inhibitors on recognition of 1359mel cells by 1359mel-specific CD8+ T cells. (C) Dose-dependent inhibition of M3-W1-B9 CD8+ T cell recognition of HEK293/B8/EBNA1-GFP target cells by protease inhibitors. Target cells were treated with different concentrations of protease inhibitors. After washes, the treated cells were cocultured with M3-W1-B9 CD8+ T cells overnight for IFN-γ release assays.
Similar articles
- CD8 T cell recognition of endogenously expressed epstein-barr virus nuclear antigen 1.
Lee SP, Brooks JM, Al-Jarrah H, Thomas WA, Haigh TA, Taylor GS, Humme S, Schepers A, Hammerschmidt W, Yates JL, Rickinson AB, Blake NW. Lee SP, et al. J Exp Med. 2004 May 17;199(10):1409-20. doi: 10.1084/jem.20040121. J Exp Med. 2004. PMID: 15148339 Free PMC article. - Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1.
Tellam J, Connolly G, Green KJ, Miles JJ, Moss DJ, Burrows SR, Khanna R. Tellam J, et al. J Exp Med. 2004 May 17;199(10):1421-31. doi: 10.1084/jem.20040191. J Exp Med. 2004. PMID: 15148340 Free PMC article. - Identification of HLA-DP3-restricted peptides from EBNA1 recognized by CD4(+) T cells.
Voo KS, Fu T, Heslop HE, Brenner MK, Rooney CM, Wang RF. Voo KS, et al. Cancer Res. 2002 Dec 15;62(24):7195-9. Cancer Res. 2002. PMID: 12499257 - Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.
Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, Khanna R, Rowe M, Wiertz EJ. Ressing ME, et al. Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review. - Avoiding proteasomal processing: the case of EBNA1.
Dantuma NP, Sharipo A, Masucci MG. Dantuma NP, et al. Curr Top Microbiol Immunol. 2002;269:23-36. doi: 10.1007/978-3-642-59421-2_2. Curr Top Microbiol Immunol. 2002. PMID: 12224511 Review.
Cited by
- HLA associations in classical Hodgkin lymphoma: EBV status matters.
Huang X, Kushekhar K, Nolte I, Kooistra W, Visser L, Bouwman I, Kouprie N, Veenstra R, van Imhoff G, Olver B, Houlston RS, Poppema S, Diepstra A, Hepkema B, van den Berg A. Huang X, et al. PLoS One. 2012;7(7):e39986. doi: 10.1371/journal.pone.0039986. Epub 2012 Jul 10. PLoS One. 2012. PMID: 22808081 Free PMC article. - Characterization of an human leucocyte antigen A2-restricted Epstein-Barr virus nuclear antigen-1-derived cytotoxic T-lymphocyte epitope.
Marescotti D, Destro F, Baldisserotto A, Marastoni M, Coppotelli G, Masucci M, Gavioli R. Marescotti D, et al. Immunology. 2010 Mar;129(3):386-95. doi: 10.1111/j.1365-2567.2009.03190.x. Epub 2009 Nov 16. Immunology. 2010. PMID: 19922423 Free PMC article. - Burkitt's lymphoma: the Rosetta Stone deciphering Epstein-Barr virus biology.
Rowe M, Kelly GL, Bell AI, Rickinson AB. Rowe M, et al. Semin Cancer Biol. 2009 Dec;19(6):377-88. doi: 10.1016/j.semcancer.2009.07.004. Epub 2009 Jul 18. Semin Cancer Biol. 2009. PMID: 19619657 Free PMC article. Review. - G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation.
Murat P, Zhong J, Lekieffre L, Cowieson NP, Clancy JL, Preiss T, Balasubramanian S, Khanna R, Tellam J. Murat P, et al. Nat Chem Biol. 2014 May;10(5):358-64. doi: 10.1038/nchembio.1479. Epub 2014 Mar 16. Nat Chem Biol. 2014. PMID: 24633353 Free PMC article. - HLA-A*02:07 is a protective allele for EBV negative and a susceptibility allele for EBV positive classical Hodgkin lymphoma in China.
Huang X, Hepkema B, Nolte I, Kushekhar K, Jongsma T, Veenstra R, Poppema S, Gao Z, Visser L, Diepstra A, van den Berg A. Huang X, et al. PLoS One. 2012;7(2):e31865. doi: 10.1371/journal.pone.0031865. Epub 2012 Feb 15. PLoS One. 2012. PMID: 22355400 Free PMC article.
References
- Kieff, E. 1995. Epstein-Barr virus–increasing evidence of a link to carcinoma. N. Engl. J. Med. 333:724–726. - PubMed
- Rickinson, A.B., and D.J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405–431. - PubMed
- Khanna, R., D.J. Moss, and S.R. Burrows. 1999. Vaccine strategies against Epstein-Barr virus-associated diseases: lessons from studies on cytotoxic T-cell-mediated immune regulation. Immunol. Rev. 170:49–64. - PubMed
- Blake, N., T. Haigh, G. Shaka'a, D. Croom-Carter, and A. Rickinson. 2000. The importance of exogenous antigen in priming the human CD8+ T cell response: lessons from the EBV nuclear antigen EBNA1. J. Immunol. 165:7078–7087. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials