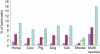Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional - PubMed (original) (raw)
Comparative Study
Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional
Kelly A Frazer et al. Genome Res. 2004 Mar.
Abstract
Cross-species DNA sequence comparison is a fundamental method for identifying biologically important elements, because functional sequences are evolutionarily conserved, wheres nonfunctional sequences drift. A recent genome-wide comparison of human and mouse DNA discovered over 200,000 conserved noncoding sequences with unknown function. Multispecies DNA comparison has been proposed as a method to prioritize these conserved noncoding sequences for functional analysis based on the hypothesis that elements present in many species are more likely to be functional than elements present in limited numbers of species. Here, we perform a comparative analysis of the single-minded 2 (SIM2) gene interval on human chromosome 21 with horse, cow, pig, dog, cat, and mouse DNA. We classify conserved sequences based on the number of mammals in which they are present, and experimentally test sequences in each class for function. As hypothesized, conserved sequences present in many mammals are frequently functional. Additionally, we demonstrate that sequences conserved in a limited number of mammals are also frequently functional. Examination of genomic deletions in chimpanzee and rhesus macaque DNA showed that several putatively functional conserved noncoding human sequences were absent in these primates. These findings suggest that functional conserved noncoding human sequences can be missing in other mammals, even closely related primate species.
Figures
Figure 1
Conserved human sequences in the 10-kb interval surrounding the first exon of SIM2 (yellow rectangle). The conserved elements are shown relative to their position in the human reference sequence (horizontal axis), and their percent conformances (vertical axis). (Position numbers) The position on human 21q (NCBI contig NT_002836). (A) Rectangles rising above the veil indicate conserved human–mouse elements identified by two methods; (blue) sequence alignment (≥100 bp and 80% identify); (gray) 21q array data (≥30 bp and 60% conformance). Red rectangles at the bottom indicate the positions of interspersed repeats, which were not tiled on the arrays, and therefore, conformance information is absent. (B) Rectangles indicate conserved human elements (≥30-nt length and ≥60% conformance) identified by comparisons of horse, cow, pig, dog, cat, and mouse DNA by hybridization to human 21q arrays.
Figure 2
Evolutionarily conserved base pairs in a 365-kb region on human chromosome 21 containing the SIM2 locus. Six species are compared with human genomic DNA for evolutionary conservation. The multispecies data set represents the composite of conserved sequences identified in all six species. The bars represent the percent of the 365,000 bp that are in elements conserved (≥30-nt length and ≥60% conformance) between humans and different numbers of the six species. Base pairs conserved only between the indicated species and humans (unique, dark blue); base pairs conserved between humans and two to five species (limited, red); base pairs conserved in all mammals analyzed (common, yellow). Pale blue bars represent the total percent of base pairs conserved. For details see Supplemental information.
Figure 3
Noncoding sequences upstream of SIM2 chosen for functional characterization. (A) Conserved sequences (c1–c10) and nonconserved sequences (n1–n5) are located within four different intervals (coordinates based on NCBI contig NT_002836) (n6 is located ∼94 kb upstream). Visualization plots (see Fig. 1) show the species in which elements c1–c10 are conserved. (B) Conserved sequences deleted in the chimpanzee and rhesus macaque genomes. Comparison of syntenic human (H), gorilla (G), chimpanzee (C), and rhesus macaque (R) long-range PCR (LR–PCR) products by gel electrophoresis shows the deletion of interval 1 conserved sequences in chimpanzee genomic DNA and the deletion of interval 3 conserved sequences in rhesus macaque genomic DNA (yellow arrows). The visualization plots, generated by hybridization of the primate LR–PCR products to the human 21q arrays, indicate the positions of the human sequences (highlighted in yellow in A and B) deleted in the chimpanzee and rhesus macaque genomes. The deletions correspond to the drop in percent conformance, plotted on the vertical scale relative to the position in the human reference sequence. Green horizontal lines at the top of the plots indicate the positions of the LR–PCR products.
Figure 4
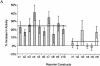
Functional characterization of noncoding sequences. (A) Expression values of 16 independent luciferase reporter constructs as assayed by transfection analysis; (filled bars) conserved noncoding sequences; (open bars) nonconserved noncoding sequences inserted in front of the SIM2 promoter (see Fig. 3A). Luciferase expression values, normalized against β-galactosidase expression values, are averages of 12 individual experiments (each construct was analyzed in triplicate on four different days) and are expressed as the percent increase in activity over the control (the luciferase reporter construct containing only the SIM2 promoter). The means across conserved and nonconserved sequences (dotted lines) were significantly different (P ∼ 0.0047, two sample _t_-test). Error bars, one SD. (B) Electrophoretic mobility-shift patterns of noncoding DNA fragments. (Lane 1, highlighted) DNA fragment alone; (lane 2) DNA fragment incubated with nuclear extract. (Red arrows) Band-shift indicating DNA–protein binding. Conserved elements c4 and c7 were too long for gel-shift analysis.
Figure 4
Functional characterization of noncoding sequences. (A) Expression values of 16 independent luciferase reporter constructs as assayed by transfection analysis; (filled bars) conserved noncoding sequences; (open bars) nonconserved noncoding sequences inserted in front of the SIM2 promoter (see Fig. 3A). Luciferase expression values, normalized against β-galactosidase expression values, are averages of 12 individual experiments (each construct was analyzed in triplicate on four different days) and are expressed as the percent increase in activity over the control (the luciferase reporter construct containing only the SIM2 promoter). The means across conserved and nonconserved sequences (dotted lines) were significantly different (P ∼ 0.0047, two sample _t_-test). Error bars, one SD. (B) Electrophoretic mobility-shift patterns of noncoding DNA fragments. (Lane 1, highlighted) DNA fragment alone; (lane 2) DNA fragment incubated with nuclear extract. (Red arrows) Band-shift indicating DNA–protein binding. Conserved elements c4 and c7 were too long for gel-shift analysis.
Similar articles
- Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment.
Dermitzakis ET, Kirkness E, Schwarz S, Birney E, Reymond A, Antonarakis SE. Dermitzakis ET, et al. Genome Res. 2004 May;14(5):852-9. doi: 10.1101/gr.1934904. Epub 2004 Apr 12. Genome Res. 2004. PMID: 15078857 Free PMC article. - Parallel construction of orthologous sequence-ready clone contig maps in multiple species.
Thomas JW, Prasad AB, Summers TJ, Lee-Lin SQ, Maduro VV, Idol JR, Ryan JF, Thomas PJ, McDowell JC, Green ED. Thomas JW, et al. Genome Res. 2002 Aug;12(8):1277-85. doi: 10.1101/gr.283202. Genome Res. 2002. PMID: 12176935 Free PMC article. - Accelerated evolution of conserved noncoding sequences in humans.
Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Prabhakar S, et al. Science. 2006 Nov 3;314(5800):786. doi: 10.1126/science.1130738. Science. 2006. PMID: 17082449 - Conserved non-genic sequences - an unexpected feature of mammalian genomes.
Dermitzakis ET, Reymond A, Antonarakis SE. Dermitzakis ET, et al. Nat Rev Genet. 2005 Feb;6(2):151-7. doi: 10.1038/nrg1527. Nat Rev Genet. 2005. PMID: 15716910 Review. - Bioinformatics for the 'bench biologist': how to find regulatory regions in genomic DNA.
Nardone J, Lee DU, Ansel KM, Rao A. Nardone J, et al. Nat Immunol. 2004 Aug;5(8):768-74. doi: 10.1038/ni0804-768. Nat Immunol. 2004. PMID: 15282556 Review.
Cited by
- Conserved Noncoding Elements Evolve Around the Same Genes Throughout Metazoan Evolution.
Gonzalez P, Hauck QC, Baxevanis AD. Gonzalez P, et al. Genome Biol Evol. 2024 Apr 2;16(4):evae052. doi: 10.1093/gbe/evae052. Genome Biol Evol. 2024. PMID: 38502060 Free PMC article. - Comparative analysis of the myoglobin gene in whales and humans reveals evolutionary changes in regulatory elements and expression levels.
Sackerson C, Garcia V, Medina N, Maldonado J, Daly J, Cartwright R. Sackerson C, et al. PLoS One. 2023 Aug 29;18(8):e0284834. doi: 10.1371/journal.pone.0284834. eCollection 2023. PLoS One. 2023. PMID: 37643191 Free PMC article. - Toward a comprehensive catalog of regulatory elements.
Fan K, Pfister E, Weng Z. Fan K, et al. Hum Genet. 2023 Aug;142(8):1091-1111. doi: 10.1007/s00439-023-02519-3. Epub 2023 Mar 19. Hum Genet. 2023. PMID: 36935423 Review. - Molecular hyperdiversity and evolution in very large populations.
Cutter AD, Jovelin R, Dey A. Cutter AD, et al. Mol Ecol. 2013 Apr;22(8):2074-95. doi: 10.1111/mec.12281. Epub 2013 Mar 18. Mol Ecol. 2013. PMID: 23506466 Free PMC article. - Genomic approaches towards finding cis-regulatory modules in animals.
Hardison RC, Taylor J. Hardison RC, et al. Nat Rev Genet. 2012 Jun 18;13(7):469-83. doi: 10.1038/nrg3242. Nat Rev Genet. 2012. PMID: 22705667 Free PMC article. Review.
References
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394. - PubMed
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578–582. - PubMed
- Ema, M., Ikegami, S., Hosoya, T., Mimura, J., Ohtani, H., Nakao, K., Inokuchi, K., Katsuki, M., and Fujii-Kuriyama, Y. 1999. Mild impairment of learning and memory in mice overexpressing the mSim2 gene located on chromosome 16: An animal model of Down's syndrome. Hum. Mol. Genet. 8: 1409–1415. - PubMed
- Fahrenkrug, S.C., Rohrer, G.A., Freking, B.A., Smith, T.P., Osoegawa, K., Shu, C.L., Catanese, J.J., and de Jong, P.J. 2001. A porcine BAC library with tenfold genome coverage: A resource for physical and genetic map integration. Mamm. Genome 12: 472–474. - PubMed
WEB SITE REFERENCES
- http://bio.cse.psu.edu/genome/hummus/; Whole Genome Human/Mouse Homology Web site.
- http://bacpac.chori.org/; BACPAC Resources Center Home Page (Children's Hospital Oakland Research Institute).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous