Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus - PubMed (original) (raw)
Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus
Eugene V Agapov et al. J Virol. 2004 Mar.
Abstract
Despite increasing characterization of pestivirus-encoded proteins, functions for nonstructural (NS) proteins NS2, NS2-3, NS4B, and NS5A have not yet been reported. Here we investigated the function of bovine viral diarrhea virus (BVDV) uncleaved NS2-3. To test whether NS2-3 has a discrete function, the uncleaved protein was specifically abolished in two ways: first by inserting a ubiquitin monomer between NS2 and NS3, and second by placing an internal ribosome entry site between the two proteins (a bicistronic genome). In both cases, complete processing of NS2-3 prevented infectious virion formation without affecting RNA replication. We tested the hypothesis that uncleaved NS2-3 was involved in morphogenesis by creating a bicistronic genome in which NS2-3 was restored in the second cistron. With this genome, both uncleaved NS2-3 expression and particle production returned. We then investigated the minimal regions of the polyprotein that could rescue an NS2-3 defect by developing a trans-complementation assay. We determined that the expression of NS4A in cis with NS2-3 markedly increased its activity, while p7 could be supplied in trans. Based on these data, we propose a model for NS2-3 action in virion morphogenesis.
Figures
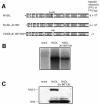
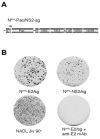
FIG. 1.
Insertion of ubiquitin between NS2 and NS3 in NADL Jiv 90− abolishes infectious particle production. (A) Schematic representations of the genomes of NADL, NADL Jiv 90−, and NADL Jiv 90−/Ubi. Specific infectivities (PFU or FFU/μg) were determined by infectious center assay and are the averages of two independent experiments. (B) RNA replication. MDBK cells were electroporated with the indicated RNA and labeled with H332PO4 in the presence of actinomycin D for 8 h starting at 16 h postelectroporation. Total cellular RNA was isolated with TRIzol, denatured, and separated on a 1% agarose gel. (C) Protein expression. Cells electroporated with the indicated RNA were lysed 10 h postelectroporation, separated by SDS-10% PAGE, Western blotted, and probed with NS3-specific polyclonal antiserum G40.
FIG. 2.
Insertion of an IRES between NS2 and NS3 in NADL Jiv 90− abolishes infectious particle production. (A) Schematic representation of the genomes of NADL, NADL Jiv 90−, subgenome (sg), Npro-E2/sg, and Npro-NS2/sg. Titers were determined by plaque or focus-forming assay. The two open reading frames of the bicistronic constructs are driven by the BVDV IRES (black line) and the EMCV IRES (dotted line). (B) RNA replication. MDBK cells were electroporated with the indicated RNAs and labeled with [3H]uridine in the presence of actinomycin D for 12 h starting at 5 h postelectroporation. Total cellular RNA was isolated with TRIzol, denatured, and separated on a 1% agarose gel. (C) Protein expression. MDBK cells electroporated with the indicated RNAs were metabolically labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation. Expression of NS3 was analyzed by immunoprecipitation with NS3-specific polyclonal antiserum G40 and separation by SDS-10% PAGE.
FIG. 3.
Bicistronic genomes can be packaged by NADL Jiv90−. MDBK cells were infected with NADL Jiv 90− at a multiplicity of infection (MOI) of 1 48 h before electroporation with bicistronic RNAs. Media were harvested at 48 h postelectroporation and used to infect naïve MDBK cells. The newly infected cells were labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation, and expression of NS3 was analyzed by immunoprecipitation with NS3-specific antiserum G40) and separation by SDS-12% PAGE. Lysates from NADL-, NADL Jiv 90−-, subgenome-, and mock-infected cells were used as controls for native protein sizes. The positions of NS3, NS2-3, and uncleaved Npro-NS3 from the subgenomic cistron are indicated.
FIG. 4.
Bicistronic genomes can package a modified defective interfering genome. (A) Schematic representation of Npro-PAC/NS2-sg, a bicistronic genome that expresses ubiquitin (Ubi, open box) and the dominant selectable marker puromycin _N_-acetyltransferase (PAC) from the first cistron and the NADL Jiv 90− NS proteins from the second cistron (p7-NS5B). Puromycin selection was used to establish a population of Npro-PAC/NS2-sg-expressing MDBK cells. (B) These cells were then electroporated with the indicated bicistronic constructs and the media were harvested 48 h postelectroporation. The supernatants were assayed for infectivity by determining their ability to transfer puromycin resistance to naïve cells. Drug-resistant foci in the newly infected cells were visualized by staining with crystal violet after 10 days of puromycin selection. Incubation of supernatants with anti-E2 antibodies abolished infectivity.
FIG.5.
Expression of NS2-3 restores virion production. (A) Schematic representation of Npro-E2/sg, Npro-E2/NS2-sg, Npro-E2/NS2-Ubi-sg, and Npro-NS2/NS2-Ubi-sg. Jiv90 (black bar) in Npro-E2/NS2-sg and ubiquitin (open box) in Npro-E2/NS2-Ubi-sg and Npro-NS2/NS2-Ubi-sg are shown. Titers of virus were determined by plaque assay or focus-forming assay. (B) Protein expression. MDBK cells electroporated with the indicated RNA were labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation. Lysates were immunoprecipitated with NS3-specific antiserum G40 and separated by SDS-10% PAGE. (C) Determination of infectious virus production. Media from cells transfected with the indicated genome were harvested after 48 h and used to infect naïve cells. Infected cells were fixed after 30 h and stained with an NS3-specific antibody (monoclonal antibody 184). Fields are magnified 10 times.
FIG. 6.
Minimal genomic regions that can rescue an uncleaved NS2-3 defect in trans. (A) Schematic representation of the genomes of Npro-NS2/sg, p7-NS2-3-4A/sg, Ubi-NS2-3-4A/sg, p7-NS2-3/sg, and p7-NS2-3(S1842A)/sg. The separate cistrons are driven by the BVDV IRES and the EMCV IRES as described for Fig. 2. The first cistron of each genome expresses the proteins shown as well as portions of the upstream and downstream open reading frames to allow correct processing (see Materials and Methods for details). Ubiquitin (open box) in Ubi-NS2-3-4A/sg and the serine-to-alanine mutation in a catalytic residue of the NS3 protease (S1842, star) in p7-NS2-3(S1842A)/sg are shown. (B) Replication of bicistronic genomes. Replication efficiencies were compared by determining NS3 levels in electroporated cells by flow cytometry (upper panel) and Western blot (lower panel). For flow cytometry, cells were fixed 30 h postelectroporation and stained with an NS3-specific antibody (monoclonal antibody 184). The percentage of electroporated cells expressing NS3 as determined by flow cytometry in two separate experiments is plotted. ND, not done. For Western blotting, cells were lysed 24 h postelectroporation and separated by SDS-10% PAGE, and the blot was probed with NS3-specific antiserum G40. The positions of NS2-3 and NS3 are indicated. β-Actin was used as a loading control. (C) Titers of virus produced by _trans_-complementation. MDBK cells were electroporated with Npro-NS2/sg alone or in combination with the indicated RNAs. Supernatants were harvested at 48 h postelectroporation and used to infect naïve cells. The appearance of NS3 in the newly infected cells was assayed at 30 h postinfection by immunofluorescence (left panel) and flow cytometry (FACS, right panel). The _trans_-complementation assay was repeated several times, and the titers were calculated by flow cytometry from two separate experiments. Flow cytometry titers were similar to those estimated by counting immunofluorescence-positive cells. Representative immunofluorescence and flow cytometry data are shown. For immunofluorescence, NS3-positive cells are red and nuclei are stained blue with Hoechst 33342. Fields are magnified 10 times. For flow cytometry, 105 cells were counted and forward scatter versus NS3 expression of the live cell subset was plotted. The NS3-positive cells are enclosed in the red gate, and the percentage of positive cells is indicated. The top row shows the supernatant assay of a representative single electroporation, Npro-NS2/sg.
FIG. 6.
Minimal genomic regions that can rescue an uncleaved NS2-3 defect in trans. (A) Schematic representation of the genomes of Npro-NS2/sg, p7-NS2-3-4A/sg, Ubi-NS2-3-4A/sg, p7-NS2-3/sg, and p7-NS2-3(S1842A)/sg. The separate cistrons are driven by the BVDV IRES and the EMCV IRES as described for Fig. 2. The first cistron of each genome expresses the proteins shown as well as portions of the upstream and downstream open reading frames to allow correct processing (see Materials and Methods for details). Ubiquitin (open box) in Ubi-NS2-3-4A/sg and the serine-to-alanine mutation in a catalytic residue of the NS3 protease (S1842, star) in p7-NS2-3(S1842A)/sg are shown. (B) Replication of bicistronic genomes. Replication efficiencies were compared by determining NS3 levels in electroporated cells by flow cytometry (upper panel) and Western blot (lower panel). For flow cytometry, cells were fixed 30 h postelectroporation and stained with an NS3-specific antibody (monoclonal antibody 184). The percentage of electroporated cells expressing NS3 as determined by flow cytometry in two separate experiments is plotted. ND, not done. For Western blotting, cells were lysed 24 h postelectroporation and separated by SDS-10% PAGE, and the blot was probed with NS3-specific antiserum G40. The positions of NS2-3 and NS3 are indicated. β-Actin was used as a loading control. (C) Titers of virus produced by _trans_-complementation. MDBK cells were electroporated with Npro-NS2/sg alone or in combination with the indicated RNAs. Supernatants were harvested at 48 h postelectroporation and used to infect naïve cells. The appearance of NS3 in the newly infected cells was assayed at 30 h postinfection by immunofluorescence (left panel) and flow cytometry (FACS, right panel). The _trans_-complementation assay was repeated several times, and the titers were calculated by flow cytometry from two separate experiments. Flow cytometry titers were similar to those estimated by counting immunofluorescence-positive cells. Representative immunofluorescence and flow cytometry data are shown. For immunofluorescence, NS3-positive cells are red and nuclei are stained blue with Hoechst 33342. Fields are magnified 10 times. For flow cytometry, 105 cells were counted and forward scatter versus NS3 expression of the live cell subset was plotted. The NS3-positive cells are enclosed in the red gate, and the percentage of positive cells is indicated. The top row shows the supernatant assay of a representative single electroporation, Npro-NS2/sg.
FIG. 7.
Flaviviridae capsid protein carboxy termini contain viral protease cleavage sites. The membrane topology of the capsid-envelope protein junction of the pestiviruses (Erns), hepatitis C virus (E1), and flaviviruses (prM) is diagrammed (top). The site of signal peptidase cleavage of the envelope proteins from capsid is shown. A second cleavage takes place in the region of the open box (arrow with star). This cleavage is mediated by the NS2B-3 proteases (flaviviruses), signal peptide peptidase (hepatitis C virus), and an unknown enzyme (pestiviruses). The exact site of cleavage in the pestiviruses and hepatitis C virus is not known. Sequences of the region of the capsid protein carboxy termini outlined by the open box above are shown in detail (bottom). Representative sequences from the three genera are shown (pestivirus, NADL; flavivirus, yellow fever virus; hepatitis C virus, genotype 1a [19]). Conserved amino acids are shown in bold. The recognition sites of the appropriate viral proteases are underlined. While none of the flavivirus protease recognition sites are absolutely conserved, they generally consist of two basic amino acids before the cleavage site and a residue with a short side chain immediately following it.
Similar articles
- Packaging defects in pestiviral NS4A can be compensated by mutations in NS2 and NS3.
Fellenberg J, Dubrau D, Isken O, Tautz N. Fellenberg J, et al. J Virol. 2023 Sep 28;97(9):e0057223. doi: 10.1128/jvi.00572-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37695056 Free PMC article. - Non-structural proteins of bovine viral diarrhea virus.
Chi S, Chen S, Jia W, He Y, Ren L, Wang X. Chi S, et al. Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review. - Characterization of the Determinants of NS2-3-Independent Virion Morphogenesis of Pestiviruses.
Klemens O, Dubrau D, Tautz N. Klemens O, et al. J Virol. 2015 Nov;89(22):11668-80. doi: 10.1128/JVI.01646-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355097 Free PMC article. - Determination of Critical Requirements for Classical Swine Fever Virus NS2-3-Independent Virion Formation.
Dubrau D, Schwindt S, Klemens O, Bischoff H, Tautz N. Dubrau D, et al. J Virol. 2019 Aug 28;93(18):e00679-19. doi: 10.1128/JVI.00679-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31292243 Free PMC article. - Molecular virology of the hepatitis C virus.
De Francesco R. De Francesco R. J Hepatol. 1999;31 Suppl 1:47-53. doi: 10.1016/s0168-8278(99)80374-2. J Hepatol. 1999. PMID: 10622560 Review.
Cited by
- Packaging defects in pestiviral NS4A can be compensated by mutations in NS2 and NS3.
Fellenberg J, Dubrau D, Isken O, Tautz N. Fellenberg J, et al. J Virol. 2023 Sep 28;97(9):e0057223. doi: 10.1128/jvi.00572-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37695056 Free PMC article. - Endogenous Viral Elements in Shrew Genomes Provide Insights into Pestivirus Ancient History.
Li Y, Bletsa M, Zisi Z, Boonen I, Gryseels S, Kafetzopoulou L, Webster JP, Catalano S, Pybus OG, Van de Perre F, Li H, Li Y, Li Y, Abramov A, Lymberakis P, Lemey P, Lequime S. Li Y, et al. Mol Biol Evol. 2022 Oct 7;39(10):msac190. doi: 10.1093/molbev/msac190. Mol Biol Evol. 2022. PMID: 36063436 Free PMC article. - Proteolytic Processing of the Coronavirus Replicase Nonstructural Protein 14 Exonuclease Is Not Required for Virus Replication but Alters RNA Synthesis and Viral Fitness.
Anderson-Daniels J, Gribble J, Denison M. Anderson-Daniels J, et al. J Virol. 2022 Aug 24;96(16):e0084122. doi: 10.1128/jvi.00841-22. Epub 2022 Aug 4. J Virol. 2022. PMID: 35924922 Free PMC article. - DNAJC14-Independent Replication of the Atypical Porcine Pestivirus.
Reuscher CM, Seitz K, Schwarz L, Geranio F, Isken O, Raigel M, Huber T, Barth S, Riedel C, Netsch A, Zimmer K, Rümenapf T, Tautz N, Lamp B. Reuscher CM, et al. J Virol. 2022 Aug 10;96(15):e0198021. doi: 10.1128/jvi.01980-21. Epub 2022 Jul 19. J Virol. 2022. PMID: 35852352 Free PMC article. - Non-structural proteins of bovine viral diarrhea virus.
Chi S, Chen S, Jia W, He Y, Ren L, Wang X. Chi S, et al. Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review.
References
- Baroth, M., M. Orlich, H. J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278:456-466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous