A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen - PubMed (original) (raw)
A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen
Hong Xu et al. EMBO J. 2004.
Abstract
The Drosophila visual system has provided a model to study phototransduction and retinal degeneration. To identify new candidate proteins that contribute to these processes, we conducted a genome-wide screen for genes expressed predominately in the eye, using DNA microarrays. This screen appeared to be comprehensive as it led to the identification of all 22 eye-enriched genes previously shown to function in phototransduction or implicated in retinal degeneration. In addition, we identified 93 eye-enriched genes whose roles have not been previously defined. One of the eye-enriched genes encoded a member of a large family of transmembrane proteins, referred to as tetraspanins. We created a null mutation in the eye-enriched tetraspanin, Sunglasses (Sun), which resulted in light-induced retinal degeneration. We found that the Sun protein was distributed primarily in lysosomes, and functioned in a long-known but poorly understood phenomenon of light-induced degradation of rhodopsin. We propose that lysosomal tetraspanins in mammalian cells may also function in the downregulation of rhodopsin and other G-protein-coupled receptors, in response to intense or prolonged agonist stimulation.
Figures
Figure 1
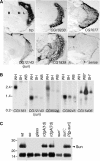
Expression patterns of sun and novel eye-enriched genes. (A) Spatial distributions of selected eye-enriched mRNAs in adult heads. In situ hybridizations to frontal sections were performed with antisense RNAs, except for the trp sense control (sense). The Flybase ID numbers are indicated. B, brain; L, lamina; M, medulla; R, retina. (B) Expression of sun and selected eye-enriched mRNAs by Northern blot analysis. Lanes contained mRNAs prepared from wt fly heads (WH), so heads (SH) and wt bodies (WB). CG15406 was predicted to be body enriched, based on the GeneChip data (Supplementary Table 3). *The order of the SH and WB samples probed with the CG15406 DNA was reversed relative to the other samples. The filters were reprobed with rp49, demonstrating that the RNA loading in each lane was similar (data not shown). (C) The Sun protein was enriched in photoreceptor cells. Anti-Sun antibodies were used to probe a Western blot containing Triton X-100 soluble proteins extracted from the indicated 1-day-old flies (except for 7-day-old rdgA). The lower 15 kDa protein of unknown identity cross-reacted with the anti-Sun antibodies. This band provided an internal loading control.
Figure 2
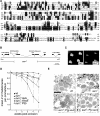
sun gene and sun 1 retinal degeneration phenotype. (A) Alignment of Sun with other tetraspanins. The putative transmembrane segments are underlined. *Residues that are ⩾80% identical among all tetraspanins. Invariant residues are boxed. (B) sun gene. The two open triangles at the left and right ends indicate the locations of the primers used to screen for deletions generated by imprecise excision of the P-element, EP(2)2425. The 1.8 kb deletion in sun 1 is indicated. (C) Rhabdomeres from 4-week-old wt and sun 1 flies observed using the optica neutralization technique (Franceschini and Kirschfeld, 1971). The flies were reared at 25°C under a 12 h light/12 h dark cycle. (D) Mean number of rhabdomeres/ommatidium as a function of age. The flies were reared at 25°C under a 12 h light/12 h dark cycle, except for the dark-reared sun 1 flies. The number of rhabdomeres was determined by the optical neutralization technique. Each data point was based on examination of ⩾90 ommatidia from ⩾8 flies. Error bars represent the SDs. (E) Retinal morphology of 4-week-old flies examined by transmission EM. Tangential sections were obtained from the distal region of the compound eye. Sections from wt, sun 1, the genomic transgene in a sun 1 background (sun 1;P[_g-sun_]) and the _myc_-sun cDNA transgene in a sun 1 background (sun 1;P[_c-sun_]) are shown.
Figure 3
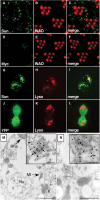
Immunolocalization of Sun. (A–F) Fly heads were embedded in LR White and 0.5 μm cross-sections of compound eyes were stained with rabbit anti-Myc and a rhabdomeral marker, rat anti-INAD. (A) Section from myc-sun transgenic fly, P[w+;_c-sun_];cn bw, stained with rabbit anti-Myc polyclonal antibodies. (B) Same section as in (A) stained with rat anti-INAD antibodies. (C) Merged images from (A, B). (D) Section from a white-eyed (cn bw) nontransgenic fly stained with anti-Myc antibodies. (E) Same section as in (D) stained with anti-INAD. (F) Merged images from (D, E). (G–I) 293 cells transfected with a plasmid encoding a YFP-Sun fusion protein. (G) Localization of YFP. (H) Same cells as in (J) stained with LysoTracker. (I) Merged images from (G, H). (J–L) 293 cells transfected with a plasmid encoding YFP. (J) Localization of YFP. (K) Same cells as (G) stained with LysoTracker. (L) Merged images from (J, K). (M, N) Immuno-EM localization of Myc-Sun and Rh1 in the cell bodies of P[_c-sun_];cn bw photoreceptor cells. Sections were stained with anti-Myc rabbit polyclonal antibodies (15 nm gold particles; several are indicated by arrows) and anti-Rh1 mouse monoclonal antibodies (5 nm gold particles, several are indicated by arrowheads). The dashed and solid boxes show lower and higher magnification views of Myc-Sun and Rh1 positive lysosomal-like vesicles (M) and MVB-like vesicles (N). The small (inset) and large-scale bars represent 0.1 and 1.0 μm, respectively. Mt, mitochondrion; Nu, nucleus; Rha, rhabdomere.
Figure 4
Defect in blue light-induced lysosomal degradation of Rh1 in sun 1. (A) Rh1 levels in wt and sun 1 flies. Three-day-old flies were either kept under ambient light (A) or stimulated for 6 h with blue light (L). Head extracts were prepared and a Western blot was probed with anti-Rh1 antibodies and 125I-labeled-anti-mouse IgG. Parallel Western blots, using the same samples, were probed with anti-TRP, anti-INAD and anti-Arr2 antibodies. (B) Quantitation of relative Rh1 levels from (A). Band intensities were determined using a phosphoimager. (C) Rh1 co-immunoprecipitated (co-IPed) with Sun in vivo. Head extracts were prepared from wt or sun 1. IPs were performed with anti-Sun antibodies or preimmune serum and a Western blot was probed with anti-Rh1 antibodies. A 1% input in lanes 1–3 corresponds to lanes 4–6, respectively. Lanes: 4, IP with anti-Sun and sun 1 extracts; 5, IP with preimmune serum and wt extracts; 6, IP with anti-Sun and wt extracts. In some but not all experiments with nonimmune serum, an ∼35 kDa band was detected. This band was not detected with anti-Sun antibodies. (D) A lysosomal inhibitor suppressed blue light-induced degradation of Rh1. Isolated wt or sun 1 ommatidia were maintained in the dark (D) or exposed to blue light for 4 h (L). Prior to the light treatment, some of the ommatidia were transferred to media containing either a lysosomal inhibitor (LI; 10 μg/μl leupeptin) or a proteasomal inhibitor (PI; 10 μM lactacystin). The Western blot was probed with anti-Rh1 antibodies and 125I-labeled-anti-mouse IgG. Parallel Western blots containing the same samples were probed with antibodies that recognize the retinal-enriched proteins INAD and TRP. (E) Quantitation of the relative Rh1 levels from (D). The relative intensities of the bands were determined using a phosphoimager.
Figure 5
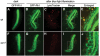
Light-dependent translocation of Rh1 in wt but not sun 1 isolated ommatidia. (A–E) Ommatidia from wt expressing a GFP-Rh1 transgene. (F–J) Ommatidia from sun 1 expressing a GFP-Rh1 transgene. (A, F) GFP-Rh1 staining (green) in ommatidia maintained in the dark. (B, G) Ommatidia incubated in media containing LysoTracker and exposed to blue light for 1.5 h. The GFP-Rh1 expression is shown (green). (C, H) LysoTracker staining (red) in the same ommatidia shown in (B) and (G), respectively. (D, I) Merge of staining shown in (B, C) and (G, H), respectively. (E, J) Higher magnifications of regions indicated by boxes in (D, I), respectively.
Similar articles
- Phototransduction and retinal degeneration in Drosophila.
Wang T, Montell C. Wang T, et al. Pflugers Arch. 2007 Aug;454(5):821-47. doi: 10.1007/s00424-007-0251-1. Epub 2007 May 9. Pflugers Arch. 2007. PMID: 17487503 Review. - Autophagy-dependent rhodopsin degradation prevents retinal degeneration in Drosophila.
Midorikawa R, Yamamoto-Hino M, Awano W, Hinohara Y, Suzuki E, Ueda R, Goto S. Midorikawa R, et al. J Neurosci. 2010 Aug 11;30(32):10703-19. doi: 10.1523/JNEUROSCI.2061-10.2010. J Neurosci. 2010. PMID: 20702701 Free PMC article. - Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation.
Kang MJ, Ryoo HD. Kang MJ, et al. Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17043-8. doi: 10.1073/pnas.0905566106. Epub 2009 Sep 23. Proc Natl Acad Sci U S A. 2009. PMID: 19805114 Free PMC article. - Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration.
Chinchore Y, Mitra A, Dolph PJ. Chinchore Y, et al. PLoS Genet. 2009 Feb;5(2):e1000377. doi: 10.1371/journal.pgen.1000377. Epub 2009 Feb 13. PLoS Genet. 2009. PMID: 19214218 Free PMC article.
Cited by
- Ih channels control feedback regulation from amacrine cells to photoreceptors.
Hu W, Wang T, Wang X, Han J. Hu W, et al. PLoS Biol. 2015 Apr 1;13(4):e1002115. doi: 10.1371/journal.pbio.1002115. eCollection 2015 Apr. PLoS Biol. 2015. PMID: 25831426 Free PMC article. - Phototransduction and retinal degeneration in Drosophila.
Wang T, Montell C. Wang T, et al. Pflugers Arch. 2007 Aug;454(5):821-47. doi: 10.1007/s00424-007-0251-1. Epub 2007 May 9. Pflugers Arch. 2007. PMID: 17487503 Review. - Drosophila visual transduction.
Montell C. Montell C. Trends Neurosci. 2012 Jun;35(6):356-63. doi: 10.1016/j.tins.2012.03.004. Epub 2012 Apr 10. Trends Neurosci. 2012. PMID: 22498302 Free PMC article. Review. - Molecular components affecting ocular carotenoid and retinoid homeostasis.
von Lintig J, Moon J, Babino D. von Lintig J, et al. Prog Retin Eye Res. 2021 Jan;80:100864. doi: 10.1016/j.preteyeres.2020.100864. Epub 2020 Apr 25. Prog Retin Eye Res. 2021. PMID: 32339666 Free PMC article. Review. - The cuticular nature of corneal lenses in Drosophila melanogaster.
Stahl AL, Charlton-Perkins M, Buschbeck EK, Cook TA. Stahl AL, et al. Dev Genes Evol. 2017 Jul;227(4):271-278. doi: 10.1007/s00427-017-0582-7. Epub 2017 May 5. Dev Genes Evol. 2017. PMID: 28477155 Free PMC article.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor Miklos GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Deitz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC (2000) The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 - PubMed
- Alloway PG, Howard L, Dolph PJ (2000) The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron 28: 129–138 - PubMed
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP (2002) Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275 - PubMed
- Bascom RA, Manara S, Collins L, Molday RS, Kalnins VI, McInnes RR (1992) Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron 8: 1171–1184 - PubMed
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL (2001) Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107: 579–589 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA92190/CA/NCI NIH HHS/United States
- R01 CA092190/CA/NCI NIH HHS/United States
- R21 CA94393/CA/NCI NIH HHS/United States
- R21 CA094393/CA/NCI NIH HHS/United States
- R01 EY008117/EY/NEI NIH HHS/United States
- EY08117/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases