A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein-coupled alpha-factor receptor - PubMed (original) (raw)
A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein-coupled alpha-factor receptor
Jennifer C Lin et al. Mol Cell Biol. 2004 Mar.
Abstract
The alpha-factor receptor (Ste2p) that promotes mating in Saccharomyces cerevisiae is similar to other G protein-coupled receptors (GPCRs) in that it contains seven transmembrane domains. Previous studies suggested that the extracellular ends of the transmembrane domains are important for Ste2p function, so a systematic scanning mutagenesis was carried out in which 46 residues near the ends of transmembrane domains 1, 2, 3, 4, and 7 were replaced with cysteine. These mutants complement mutations constructed previously near the ends of transmembrane domains 5 and 6 to analyze all the extracellular ends. Eight new mutants created in this study were partially defective in signaling (V45C, N46C, T50C, A52C, L102C, N105C, L277C, and A281C). Treatment with 2-([biotinoyl] amino) ethyl methanethiosulfonate, a thiol-specific reagent that reacts with accessible cysteine residues but not membrane-embedded cysteines, identified a drop in the level of reactivity over a consecutive series of residues that was inferred to be the membrane boundary. An unusual prolonged zone of intermediate reactivity near the extracellular end of transmembrane domain 2 suggests that this region may adopt a special structure. Interestingly, residues implicated in ligand binding were mainly accessible, whereas residues involved in the subsequent step of promoting receptor activation were mainly inaccessible. These results define a receptor microdomain that provides an important framework for interpreting the mechanisms by which functionally important residues contribute to ligand binding and activation of Ste2p and other GPCRs.
Figures
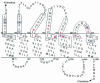
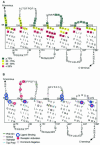
FIG. 1.
Residues targeted for Cys substitution mutagenesis near the ends of TMDs in Ste2p. Residues with Cys substituted as part of this study are boxed with solid lines. The residues boxed with a dashed line were mutated to Cys in a previous study (29). The shading indicates Cys substitution mutations that caused a significant decrease in receptor function. Red indicates residues affected by dominant-negative mutations (9, 25, 55). This topographical representation of Ste2p was the working model for the TMDs used at the start of these studies.
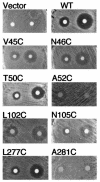
FIG. 2.
Cys substitution mutants defective in undergoing cell division arrest in response to α-factor. Yeast strain yLG123 carrying the wild-type receptor plasmid or versions containing the indicated Cys substitution mutants were assayed for the ability to undergo cell division arrest in response to α-factor. Cell division arrest leads to the formation of a zone of growth inhibition (halo) surrounding a filter disk containing α-factor (200 or 1,200 ng) applied to a lawn of cells on the surface of an agar plate.
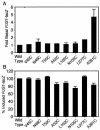
FIG. 3.
Basal and α-factor-induced levels of the pheromone-responsive FUS1-lacZ reporter gene. Shown are PMY1 cells carrying either wild-type STE2 or the indicated Cys substitution mutants on a plasmid in the absence (A) or the presence (B) of 10−6 M α-factor for 2 h. FUS1-lacZ reporter gene activity was determined by assaying β-galactosidase activity as described in Materials and Methods, and the values were normalized to the basal or maximal induced value of the wild-type cells as indicated. The results represent the averages of two to four independent experiments, each done in duplicate. The error bars represent standard deviations. A t test indicated significant differences between the basal levels of the wild type and A281C (P < 0.0001) and between the induced levels of the wild type and A52C (P < 0.005), L102C (P < 0.005), N105C (P < 0.001), and A281C (P < 0.01).
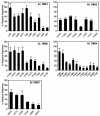
FIG. 4.
Reactivities of Cys residues with MTSEA-biotin. Accessibility assays were carried out by treating membrane fractions from the indicated Cys substitution mutants with MTSEA-biotin. The proteins were solubilized in detergent buffer, and then the biotin-labeled receptors were collected on streptavidin beads. The degree of receptor biotinylation was quantified on Western immunoblots and is reported as the percentage of labeling relative to the T199C receptors. In each experiment, control samples confirmed that the wild-type receptors lacking Cys residues did not react significantly and that the T199C receptors, which contain a Cys in the second extracellular loop, reacted well with MTSEA-biotin (38%). This level of biotinylation is likely to be an underestimate, since stringent washing conditions were used to prevent nonspecific sticking to the streptavidin beads. The results represent the averages of at least three independent experiments plus standard errors. Analysis of variance carried out using the ANOVA program (GraphPad Software, San Diego, Calif.) indicated that the means differed significantly (P < 0.01) and that there was a significant trend of decreasing accessibility for TMD1, -3, -4, and -7 (P < 0.001).
FIG. 5.
Surface accessibility maps of the α-factor receptor. (A) Reactivities of Cys residues substituted into the α-factor receptor are color-coded as indicated. This topology map was adjusted to place the membrane boundary at the start of the drop in reactivity to MTSEA-biotin. The dotted lines indicate the positions of previous predictions for the TMD boundaries, as shown in Fig. 1. (B) Comparison of topology predictions by the indicated computer programs. The bars represent the ends of the TMDs as predicted by the programs that were accessed at the ExPASy molecular biology server (
). Sites of Cys substitutions that altered the binding affinity are circled in blue, as are positions 47 and 48, which were implicated previously in binding α-factor using different approaches (27). Shaded in red are the sites of Cys substitutions that prevented maximal signaling activity in this study and also previously studied mutations at positions 102, 105, 108, and 111 (1). Residues affected by dominant-negative mutations are shaded gray (9, 25, 55).
FIG. 6.
Model for spatial relationshipsof residues implicated in ligand binding and activation of Ste2p.(A) Three-dimensional model of the extracellular ends of the TMDs based on the crystal structure of rhodopsin (7). The MTSEA-biotin accessibility data were used to map Ste2p residues near the ends of the TMDs onto the corresponding residues in rhodopsin. The side chains colored blue are implicated in binding α-factor; those colored red are implicated in receptor activation. The residues include those described in this study and previous studies, as explained in the legend to Fig. 5 (7 , 27, 29). (B) Schematic representation of TMD ends with Ste2p residue numbers labeled. A prediction for the orientation of α-factor in the binding domain is shown as a dashed green line. This prediction for how α-factor docks with the receptor was inferred from functional data and cross-linking experiments that indicate that Phe204 may interact with the C terminus of α-factor and Tyr266 may interact with the N terminus (9). Since α-factor contains a β-bend structure around a central Pro-Gly bond (7), it was proposed to dock onto a model of the receptor as viewed from the extracellular side of the plasma membrane.
Similar articles
- Aromatic residues at the extracellular ends of transmembrane domains 5 and 6 promote ligand activation of the G protein-coupled alpha-factor receptor.
Lin JC, Parrish W, Eilers M, Smith SO, Konopka JB. Lin JC, et al. Biochemistry. 2003 Jan 21;42(2):293-301. doi: 10.1021/bi026766o. Biochemistry. 2003. PMID: 12525156 - Changes in conformation at the cytoplasmic ends of the fifth and sixth transmembrane helices of a yeast G protein-coupled receptor in response to ligand binding.
Umanah GK, Huang LY, Maccarone JM, Naider F, Becker JM. Umanah GK, et al. Biochemistry. 2011 Aug 16;50(32):6841-54. doi: 10.1021/bi200254h. Epub 2011 Jul 12. Biochemistry. 2011. PMID: 21728340 Free PMC article. - Comparison of Experimental Approaches Used to Determine the Structure and Function of the Class D G Protein-Coupled Yeast α-Factor Receptor.
Dumont ME, Konopka JB. Dumont ME, et al. Biomolecules. 2022 May 30;12(6):761. doi: 10.3390/biom12060761. Biomolecules. 2022. PMID: 35740886 Free PMC article. Review.
Cited by
- The N-terminus of the yeast G protein-coupled receptor Ste2p plays critical roles in surface expression, signaling, and negative regulation.
Uddin MS, Hauser M, Naider F, Becker JM. Uddin MS, et al. Biochim Biophys Acta. 2016 Apr;1858(4):715-24. doi: 10.1016/j.bbamem.2015.12.017. Epub 2015 Dec 17. Biochim Biophys Acta. 2016. PMID: 26707753 Free PMC article. - Comparison of class A and D G protein-coupled receptors: common features in structure and activation.
Eilers M, Hornak V, Smith SO, Konopka JB. Eilers M, et al. Biochemistry. 2005 Jun 28;44(25):8959-75. doi: 10.1021/bi047316u. Biochemistry. 2005. PMID: 15966721 Free PMC article. - Studying structure and functions of cell membranes by single molecule biophysical techniques.
Zhang Q, Li S, Yang Y, Shan Y, Wang H. Zhang Q, et al. Biophys Rep. 2021 Oct 31;7(5):384-398. doi: 10.52601/bpr.2021.210018. Biophys Rep. 2021. PMID: 37288104 Free PMC article. - Comparative NMR analysis of an 80-residue G protein-coupled receptor fragment in two membrane mimetic environments.
Cohen LS, Arshava B, Neumoin A, Becker JM, Güntert P, Zerbe O, Naider F. Cohen LS, et al. Biochim Biophys Acta. 2011 Nov;1808(11):2674-84. doi: 10.1016/j.bbamem.2011.07.011. Epub 2011 Jul 23. Biochim Biophys Acta. 2011. PMID: 21791199 Free PMC article.
References
- Akal-Strader, A., S. Khare, D. Xu, F. Naider, and J. M. Becker. 2002. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J. Biol. Chem. 277:30581-30590. - PubMed
- Ballesteros, J. A., L. Shi, and J. A. Javitch. 2001. Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol. Pharmacol. 60:1-19. - PubMed
- Baranski, T. J., P. Herzmark, O. Lichtarge, B. O. Gerber, J. Trueheart, E. C. Meng, T. Iiri, S. P. Sheikh, and H. R. Bourne. 1999. C5a receptor activation. Genetic identification of critical residues in four transmembrane helices. J. Biol. Chem. 274:15757-15765. - PubMed
- Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. - PubMed
- Bourne, H. R. 1997. How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 9:134-142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases