Bordetella species are distinguished by patterns of substantial gene loss and host adaptation - PubMed (original) (raw)
Comparative Study
Bordetella species are distinguished by patterns of substantial gene loss and host adaptation
C A Cummings et al. J Bacteriol. 2004 Mar.
Abstract
Pathogens of the bacterial genus Bordetella cause respiratory disease in humans and animals. Although virulence and host specificity vary across the genus, the genetic determinants of this diversity remain unidentified. To identify genes that may underlie key phenotypic differences between these species and clarify their evolutionary relationships, we performed a comparative analysis of genome content in 42 Bordetella strains by hybridization of genomic DNA to a microarray representing the genomes of three Bordetella species and by subtractive hybridization. Here we show that B. pertussis and B. parapertussis are predominantly differentiated from B. bronchiseptica by large, species-specific regions of difference, many of which encode or direct synthesis of surface structures, including lipopolysaccharide O antigen, which may be important determinants of host specificity. The species also exhibit sequence diversity at a number of surface protein-encoding loci, including the fimbrial major subunit gene, fim2. Gene loss, rather than gene acquisition, accompanied by the proliferation of transposons, has played a fundamental role in the evolution of the pathogenic bordetellae and may represent a conserved evolutionary mechanism among other groups of microbial pathogens.
Figures
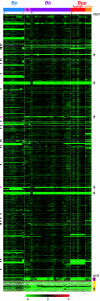
FIG.1.
CGH of 42 Bordetella strains. Each column represents a strain, ordered and color-coded according to the tree topology shown in Fig. 2. The three sequenced strains are marked by asterisks. B. pertussis strain 18323 is indicated by the dagger. Each row represents a microarray probe, arranged in RB50 gene order from “start” to “end,” with RB50 genome coordinates listed on the left. φ indicates clusters of phage genes. Arrowheads indicate regions of >5 kb not represented by a probe. Magenta bar, duplicated sequences in RB50. The chromosomal locations of the largest of these duplications, a degenerate prophage, are marked by magenta arrowheads. Yellow bar, genes not present in the RB50 sequence, arranged in Tohama I gene order. The values below the color bar at the bottom refer to log2(test/reference intensities). Missing data are gray. Bp, B. pertussis; Bb, B. bronchiseptica; Bpp, B. parapertussis.
FIG. 2.
Phylogeny of Bordetella strains based on CGH data. Phylogenetic associations were inferred using a maximum-parsimony algorithm, and confidence intervals were determined from 100 bootstrapped datasets. The scale bar represents 100 evolutionary events (steps), and bootstrap values of ≥50% are indicated (19). The presence of Bordetella IS elements, as detected by CGH, is indicated for each strain: blue boxes, IS_481_; yellow, IS_1002_; and green, IS1663. The presence of IS_1001_ (magenta), which was not represented on the array, is taken from reference . In that study, the presence of IS_1001_ was not determined for B. pertussis, excepting strains Tohama I and 18323. In parentheses following each strain number are the strain's host and alias. Sequenced strains are shaded. The diamond indicates the B. bronchiseptica clade that may be associated with B. pertussis. The inset shows an overview tree of an equally likely phylogeny, created by collapsing all branches supported by bootstrap values of ≥50.
FIG. 3.
Gene content variation and structural differences in the LPS O-antigen biosynthesis region. CGH data for the O-antigen biosynthesis region in 42 Bordetella strains are shown, displayed as in Fig. 1, with strain groups color coded according to the tree topology shown in Fig. 2. Some ORFs are represented by multiple probes. Coding sequence designations, gene names, and annotations are included. The color scale is as in Fig. 1.
FIG. 4.
Gene content variation and sequence polymorphism in fimbriae genes. Strain groups are color coded according to the tree topology shown in Fig. 2. For each probe, the sequenced strain from which it was amplified is listed, as well as the percent sequence identity of the probe to each of the three sequenced genomes. The color scale and symbols are as in Fig. 1.
Similar articles
- Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens.
Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, Ahuja U, Liu M, Miller JF, Sebaihia M, Bentley SD, Parkhill J, Harvill ET. Park J, et al. BMC Genomics. 2012 Oct 10;13:545. doi: 10.1186/1471-2164-13-545. BMC Genomics. 2012. PMID: 23051057 Free PMC article. - Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species.
Linz B, Ivanov YV, Preston A, Brinkac L, Parkhill J, Kim M, Harris SR, Goodfield LL, Fry NK, Gorringe AR, Nicholson TL, Register KB, Losada L, Harvill ET. Linz B, et al. BMC Genomics. 2016 Sep 30;17(1):767. doi: 10.1186/s12864-016-3112-5. BMC Genomics. 2016. PMID: 27716057 Free PMC article. - Isolation of a putative fimbrial adhesin from Bordetella pertussis and the identification of its gene.
Willems RJ, Geuijen C, van der Heide HG, Matheson M, Robinson A, Versluis LF, Ebberink R, Theelen J, Mooi FR. Willems RJ, et al. Mol Microbiol. 1993 Aug;9(3):623-34. doi: 10.1111/j.1365-2958.1993.tb01722.x. Mol Microbiol. 1993. PMID: 8105363 - Genetics of pertussis toxin.
Gross R, Aricò B, Rappuoli R. Gross R, et al. Mol Microbiol. 1989 Jan;3(1):119-24. doi: 10.1111/j.1365-2958.1989.tb00111.x. Mol Microbiol. 1989. PMID: 2654538 Review. - The bordetellae: lessons from genomics.
Preston A, Parkhill J, Maskell DJ. Preston A, et al. Nat Rev Microbiol. 2004 May;2(5):379-90. doi: 10.1038/nrmicro886. Nat Rev Microbiol. 2004. PMID: 15100691 Review. No abstract available.
Cited by
- A Unique Reverse Adaptation Mechanism Assists Bordetella pertussis in Resistance to Both Scarcity and Toxicity of Manganese.
Čapek J, Procházková I, Matoušek T, Hot D, Večerek B. Čapek J, et al. mBio. 2021 Oct 26;12(5):e0190221. doi: 10.1128/mBio.01902-21. Epub 2021 Oct 26. mBio. 2021. PMID: 34700381 Free PMC article. - Genetic Diversity of Clinical Bordetella Pertussis ST2 Strains in comparison with Vaccine Reference Strains of India.
Sharma NC, Anandan S, Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Vasudevan K, Kumar D, Gupta SK, Sangal L, Veeraraghavan B. Sharma NC, et al. J Genomics. 2021 Sep 3;9:38-42. doi: 10.7150/jgen.58823. eCollection 2021. J Genomics. 2021. PMID: 34527084 Free PMC article. - Comparative Phosphoproteomics of Classical Bordetellae Elucidates the Potential Role of Serine, Threonine and Tyrosine Phosphorylation in Bordetella Biology and Virulence.
Luu LDW, Zhong L, Kaur S, Raftery MJ, Lan R. Luu LDW, et al. Front Cell Infect Microbiol. 2021 Apr 13;11:660280. doi: 10.3389/fcimb.2021.660280. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33928046 Free PMC article. - Comparative Omics Analysis of Historic and Recent Isolates of Bordetella pertussis and Effects of Genome Rearrangements on Evolution.
Dienstbier A, Amman F, Petráčková D, Štipl D, Čapek J, Zavadilová J, Fabiánová K, Držmíšek J, Kumar D, Wildung M, Pouchnik D, Večerek B. Dienstbier A, et al. Emerg Infect Dis. 2021 Jan;27(1):57-68. doi: 10.3201/eid2701.191541. Emerg Infect Dis. 2021. PMID: 33350934 Free PMC article. - Bordetella Type III Secretion Injectosome and Effector Proteins.
Kamanova J. Kamanova J. Front Cell Infect Microbiol. 2020 Sep 4;10:466. doi: 10.3389/fcimb.2020.00466. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33014891 Free PMC article. Review.
References
- Andersson, J. O., and S. G. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. - PubMed
- Arico, B., R. Gross, J. Smida, and R. Rappuoli. 1987. Evolutionary relationships in the genus Bordetella. Mol. Microbiol. 1:301-308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI039587/AI/NIAID NIH HHS/United States
- R03 AI054970/AI/NIAID NIH HHS/United States
- R21 AI057188/AI/NIAID NIH HHS/United States
- R03 AI547970/AI/NIAID NIH HHS/United States
- R33 AI057188/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources