A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements - PubMed (original) (raw)
A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements
Allen S Yang et al. Nucleic Acids Res. 2004.
Abstract
We report a method for studying global DNA methylation based on using bisulfite treatment of DNA and simultaneous PCR of multiple DNA repetitive elements, such as Alu elements and long interspersed nucleotide elements (LINE). The PCR product, which represents a pool of approximately 15 000 genomic loci, could be used for direct sequencing, selective restriction digestion or pyrosequencing, in order to quantitate DNA methylation. By restriction digestion or pyrosequencing, the assay was reproducible with a standard deviation of only 2% between assays. Using this method we found that almost two-thirds of the CpG methylation sites in Alu elements are mutated, but of the remaining methylation target sites, 87% were methylated. Due to the heavy methylation of repetitive elements, this assay was especially useful in detecting decreases in DNA methylation, and this assay was validated by examining cell lines treated with the methylation inhibitor 5-aza-2'deoxycytidine (DAC), where we found a 1-16% decrease in Alu element and 18-60% LINE methylation within 3 days of treatment. This method can be used as a surrogate marker of genome-wide methylation changes. In addition, it is less labor intensive and requires less DNA than previous methods of assessing global DNA methylation.
Figures
Figure 1
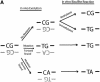
Direct DNA sequencing of bisulfite repetitive element PCR of Alu elements. (A) Schematic of possible fates of Alu element CpG methylation sites. Due to the mutation of CpG sites via spontaneous deamination of 5-methylcytosine to T during evolution, a CpG site can be changed into a TpG or CpA dinucleotide. Neither TpG nor CpA are targets for methylation. Following bisulfite treatment, a methylated CpG will remain CpG. However, an unmethylated CpG will give rise to TpG, which is indistinguishable from a deamination mutation of the forward strand. Thus, three possibilities arise for our sequencing data: CpG that represents a methylated CpG site, TpG that represents either an unmethylated CpG site or a mutation of the forward strand, and TpA that represents a mutation of the reverse strand followed by conversion of the unmethylated C to T by bisulfite. (B) Sequencing data of bisulfite repetitive element PCR of Alu elements. Genomic DNA was isolated from peripheral human blood and bisulfite treated. Alu element PCR was performed and the PCR product was cloned, and 15 clones were sequenced. There were 12 potential CpG methylation sites per clone for a total of 180 potential methylation sites sequenced. Black circles represent methylated CpG sites (66/180 = 36.7%). ‘T’ represents TpG sites that were either unmethylated or mutated (53/180 = 24.4%). ‘A’ represents TpA sites that were mutated (41/180 = 22.8%). ‘X’ represents other mutations (20/180 = 11.1%). The CpG sites used for pyrosequencing (‘PS’) and COBRA (‘MboI’) are indicated.
Figure 1
Direct DNA sequencing of bisulfite repetitive element PCR of Alu elements. (A) Schematic of possible fates of Alu element CpG methylation sites. Due to the mutation of CpG sites via spontaneous deamination of 5-methylcytosine to T during evolution, a CpG site can be changed into a TpG or CpA dinucleotide. Neither TpG nor CpA are targets for methylation. Following bisulfite treatment, a methylated CpG will remain CpG. However, an unmethylated CpG will give rise to TpG, which is indistinguishable from a deamination mutation of the forward strand. Thus, three possibilities arise for our sequencing data: CpG that represents a methylated CpG site, TpG that represents either an unmethylated CpG site or a mutation of the forward strand, and TpA that represents a mutation of the reverse strand followed by conversion of the unmethylated C to T by bisulfite. (B) Sequencing data of bisulfite repetitive element PCR of Alu elements. Genomic DNA was isolated from peripheral human blood and bisulfite treated. Alu element PCR was performed and the PCR product was cloned, and 15 clones were sequenced. There were 12 potential CpG methylation sites per clone for a total of 180 potential methylation sites sequenced. Black circles represent methylated CpG sites (66/180 = 36.7%). ‘T’ represents TpG sites that were either unmethylated or mutated (53/180 = 24.4%). ‘A’ represents TpA sites that were mutated (41/180 = 22.8%). ‘X’ represents other mutations (20/180 = 11.1%). The CpG sites used for pyrosequencing (‘PS’) and COBRA (‘MboI’) are indicated.
Figure 2
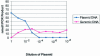
Calculation of the number of Alu elements being assessed by bisulfite Alu PCR. Competitive PCR was performed using a fixed amount of bisulfite-treated genomic DNA and a plasmid containing a cloned Alu element fragment with an internal duplication that gives a larger PCR product. A fixed amount of bisulfite-treated genomic DNA was mixed with serial dilutions of the plasmid which gave two PCR products, one from the genomic DNA and a larger one from the plasmid. The PCR products were quantitated using a capillary electrophoresis system, the Agilent 2100 Bioanalyzer (Agilent Technologies) and plotted above. From this experiment, it is approximated that 0.5 ng of plasmid gives equivalent PCR product to 50 ng of bisulfite-treated genomic DNA.
Figure 3
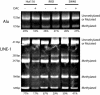
Quantitation of DNA methylation in cell lines treated with DAC using the bisulfite repetitive element PCR technique. Hct116, RKO and SW48 cell lines were treated with DAC. Genomic DNA was isolated from DAC-treated (+) and -untreated (–) controls. The genomic DNA was treated with sodium bisulfite and a non-specific PCR was performed, which amplified a pool of Alu or LINE-1 repetitive elements. The PCR product was then digested with MboI (Alu) or HinfI (LINE-1), which only cuts repetitive elements that were originally methylated. The Alu PCR assays a single methylation site and therefore the MboI digestion will cut the PCR product from 152 to 125 and 27 bp (data not shown). The LINE-1 PCR assays the methylation of two sites and therefore HinfI can generate five possible digestion products of 285, 247, 166, 128 and 38 bp (data not shown). The digested PCR product was separated by polyacrylamide gel electrophoresis and stained with ethidium bromide. The lower cut bands represent methylated repetitive elements. The upper band represents unmethylated repetitive elements or repetitive elements in which the restriction site has been mutated. The PCR bands were quantitated and the amount of methylation is shown below each gel lane. Similar results were obtained for both the Alu element and LINE-1 assays.
Figure 4
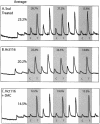
Quantitation of DNA methylation using bisulfite repetitive element PCR and pyrosequencing. (A) SssI methylase-treated DNA, (B) untreated Hct116 cell line DNA or (C) DAC-treated Hct116 cell line DNA was bisulfite treated and PCR of Alu repetitve elements was performed. The PCR product was purified and methylation was quantitated using the PSQ HS 96 Pyrosequencing System (Pyrosequencing, Inc.). The pyrogram quantitates C for methylated and T for unmethylated or mutated DNA. The shaded regions represent three tandem CpG sites quantitated in Alu elements, and the percent methylation at each site is shown above the peaks. The average methylation of the three sites is calculated on the left for each pyrogram. The maximum absolute methylation of 23.2% is calculated by SssI methylase-treated DNA (A), Hct116 cells have 20.2% methylation, and this methylation decreases to 14.5% after DAC treatment of Hct116 cells.
Similar articles
- Determining the global DNA methylation status of rat according to the identifier repetitive elements.
Kim HH, Park JH, Jeong KS, Lee S. Kim HH, et al. Electrophoresis. 2007 Nov;28(21):3854-61. doi: 10.1002/elps.200600733. Electrophoresis. 2007. PMID: 17960839 - Differential repetitive DNA methylation in multiple myeloma molecular subgroups.
Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, Motta V, Bertazzi PA, Baccarelli A, Neri A. Bollati V, et al. Carcinogenesis. 2009 Aug;30(8):1330-5. doi: 10.1093/carcin/bgp149. Epub 2009 Jun 16. Carcinogenesis. 2009. PMID: 19531770 - Droplet digital PCR for the quantification of Alu methylation status in hematological malignancies.
Orsini P, Impera L, Parciante E, Cumbo C, Minervini CF, Minervini A, Zagaria A, Anelli L, Coccaro N, Casieri P, Tota G, Brunetti C, Ricco A, Carluccio P, Specchia G, Albano F. Orsini P, et al. Diagn Pathol. 2018 Dec 22;13(1):98. doi: 10.1186/s13000-018-0777-x. Diagn Pathol. 2018. PMID: 30579366 Free PMC article. - CGGBP1 mitigates cytosine methylation at repetitive DNA sequences.
Agarwal P, Collier P, Fritz MH, Benes V, Wiklund HJ, Westermark B, Singh U. Agarwal P, et al. BMC Genomics. 2015 May 16;16(1):390. doi: 10.1186/s12864-015-1593-2. BMC Genomics. 2015. PMID: 25981527 Free PMC article. - Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer.
Baba Y, Murata A, Watanabe M, Baba H. Baba Y, et al. Surg Today. 2014 Oct;44(10):1807-16. doi: 10.1007/s00595-013-0763-6. Epub 2013 Oct 23. Surg Today. 2014. PMID: 24150097 Review.
Cited by
- Alu and LINE-1 methylation and lung function in the normative ageing study.
Lange NE, Sordillo J, Tarantini L, Bollati V, Sparrow D, Vokonas P, Zanobetti A, Schwartz J, Baccarelli A, Litonjua AA, Demeo DL. Lange NE, et al. BMJ Open. 2012 Oct 17;2(5):e001231. doi: 10.1136/bmjopen-2012-001231. Print 2012. BMJ Open. 2012. PMID: 23075571 Free PMC article. - Impact of prematurity on LINE-1 promoter methylation.
Dos Santos PVBE, Brasil AA, Milone LTV, Chalfun G, Saide SCAO, Salú MDS, de Oliveira MBG, Robaina JR, Lima-Setta F, Rodrigues-Santos G, de Magalhães-Barbosa MC, da Cunha AJLA, Prata-Barbosa A. Dos Santos PVBE, et al. Epigenomics. 2024;16(18):1253-1264. doi: 10.1080/17501911.2024.2397329. Epub 2024 Sep 19. Epigenomics. 2024. PMID: 39297700 Free PMC article. - LINE1 methylation levels in pre-diagnostic leukocyte DNA and future renal cell carcinoma risk.
Karami S, Andreotti G, Liao LM, Pfeiffer RM, Weinstein SJ, Purdue MP, Hofmann JN, Albanes D, Mannisto S, Moore LE. Karami S, et al. Epigenetics. 2015;10(4):282-92. doi: 10.1080/15592294.2015.1006505. Epub 2015 Feb 3. Epigenetics. 2015. PMID: 25647181 Free PMC article. - Cancer detection by ubiquitin carboxyl-terminal esterase L1 methylation in pancreatobiliary fluids.
Kato N, Yamamoto H, Adachi Y, Ohashi H, Taniguchi H, Suzuki H, Nakazawa M, Kaneto H, Sasaki S, Imai K, Shinomura Y. Kato N, et al. World J Gastroenterol. 2013 Mar 21;19(11):1718-27. doi: 10.3748/wjg.v19.i11.1718. World J Gastroenterol. 2013. PMID: 23555160 Free PMC article. - Genome-wide measures of DNA methylation in peripheral blood and the risk of urothelial cell carcinoma: a prospective nested case-control study.
Dugué PA, Brinkman MT, Milne RL, Wong EM, FitzGerald LM, Bassett JK, Joo JE, Jung CH, Makalic E, Schmidt DF, Park DJ, Chung J, Ta AD, Bolton DM, Lonie A, Longano A, Hopper JL, Severi G, Saffery R, English DR, Southey MC, Giles GG. Dugué PA, et al. Br J Cancer. 2016 Sep 6;115(6):664-73. doi: 10.1038/bjc.2016.237. Epub 2016 Aug 4. Br J Cancer. 2016. PMID: 27490804 Free PMC article.
References
- Bird A. (1992) The essentials of DNA methylation. Cell, 70, 5–8. - PubMed
- Jones P.A. and Takai,D. (2001) The role of DNA methylation in mammalian epigenetics. Science, 293, 1068–1070. - PubMed
- Jones P.A. and Baylin,S.B. (2002) The fundamental role of epigenetic events in cancer. Nature Rev. Genet., 3, 415–428. - PubMed
- Issa J.P. (2000) CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol., 249, 101–118. - PubMed
- Richardson B. (2003) Impact of aging on DNA methylation. Ageing Res. Rev., 2, 245–261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- P50CA100632/CA/NCI NIH HHS/United States
- P50 CA100632/CA/NCI NIH HHS/United States
- CA16672/CA/NCI NIH HHS/United States
- R33CA89837/CA/NCI NIH HHS/United States
- R33 CA089837/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources