The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations - PubMed (original) (raw)
The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations
Sharon Cantor et al. Proc Natl Acad Sci U S A. 2004.
Erratum in
- Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6834
Abstract
BACH1 is a nuclear protein that directly interacts with the highly conserved, C-terminal BRCT repeats of the tumor suppressor, BRCA1. Mutations within the BRCT repeats disrupt the interaction between BRCA1 and BACH1, lead to defects in DNA repair, and result in breast and ovarian cancer. BACH1 is necessary for efficient double-strand break repair in a manner that depends on its association with BRCA1. Moreover, some women with early-onset breast cancer and no abnormalities in either BRCA1 or BRCA2 carry germline BACH1 coding sequence changes, suggesting that abnormal BACH1 function contributes to tumor induction. Here, we show that BACH1 is both a DNA-dependent ATPase and a 5'-to-3' DNA helicase. In two patients with early-onset breast cancer who carry distinct germline BACH1 coding sequence changes, the resulting proteins are defective in helicase activity, indicating that these sequence changes disrupt protein function. These results reinforce the notion that mutant BACH1 participates in breast cancer development.
Figures
Fig. 1.
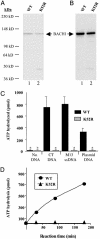
Purified BACH1 is a DNA-dependent ATPase. (A) FLAG WT and mutant (K52R) BACH1 proteins were purified by FLAG-affinity chromatography from High Five insect cells infected with the corresponding baculoviruses. The proteins were resolved in 4–12% gradient SDS-polyacrylamide gels and stained with Coomassie blue. Lane 1, 500 ng of WT BACH1; lane 2, 500 ng of K52R mutant. (B) Western blot analysis of recombinant proteins. The blot was probed with polyclonal BACH1 antiserum (E67) that recognizes the C terminus of BACH1. Lane 1, 25 ng of WT BACH1; lane 2, 25 ng of K52R mutant. (C) WT BACH1 hydrolyzes ATP in the presence of DNA. Calf thymus (CT), single-stranded (M13), and double-stranded plasmid DNA served as nucleic acid cofactors. WT and K52R mutant BACH1 proteins (600 ng each) were incubated with [γ-32P]ATP-containing reaction mixtures supplemented with 2 μg of the indicated DNA for 60 min at 37°C. ATP hydrolysis was quantitated as described in Materials and Methods. The asterisk denotes negligible ATPase activity above background. Results were calculated from experiments performed in triplicate. (D) ATP hydrolysis by BACH1 is approximately linear over time. WT and K52R mutant BACH1 (600 ng each) were incubated in the above-noted [γ-32P]ATP- and CT DNA (2 μg) containing reaction mixture for the times indicated, and the results were determined. Similar results were obtained in at least three independent experiments.
Fig. 2.
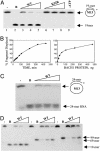
BACH1 is an ATP-dependent helicase. (A) Increasing amounts of WT and K52R mutant BACH1 were incubated with a DNA helicase substrate containing an annealed radiolabeled 19-nt oligomer (see Materials and Methods). Lane 1, annealed substrate (–); lane 2, heat-denatured substrate (B, for boiled); lanes 3–5, BACH1 (60, 180, and 450 ng, respectively); lanes 6–8, K52R BACH1 (200, 400, and 600 ng, respectively); lane 9, WT with no ATP. (B) BACH1 unwinds DNA in a time- and dose-dependent manner. BACH1 protein (150 ng) was incubated with the 19-mer helicase substrate for the indicated times. Independently, increasing amounts of BACH1 (15, 30, 60, 120, 240, and 480 ng) were incubated with substrate for 30 min. (C) Increasing amounts of BACH1 (60, 180, and 450 ng) were incubated with a RNA:DNA helicase substrate and helicase activity was measured. (D) Increasing quantities of BACH1 (60, 180, and 450 ng) were incubated with DNA helicase substrates of increasing partial duplex length, as indicated. In all cases, reaction products were resolved in an 8% native polyacrylamide gel containing 15% glycerol. Results were quantitated by using a Molecular Dynamics STORM PhosphorImager.
Fig. 3.
BACH1 preferentially unwinds DNA in the 5′-to-3′ direction. (A) Scheme for helicase directionality assay. A single-stranded 92-mer oligodeoxynucleotide was annealed to M13 DNA and cleaved with _Sal_I, and all available 3′ ends were radiolabeled by using Klenow polymerase to yield a partially double-stranded substrate comprising 38-mer and 55-mer oligonucleotides annealed to linear M13 DNA. (B) Autoradiogram of an 8% polyacrylamide gel of the products generated by incubating the substrate depicted in A with FLAG-BACH1. Lane 1, substrate alone; lane 2, denatured substrate (B); lane 3, 5 ng of UvrD protein as a 3′-to-5′ helicase control; lanes 4–6, substrate plus 60, 150, and 450 ng of FLAG-BACH1.
Fig. 4.
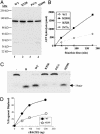
Germ-line sequence changes in BACH1 disrupt BACH1 helicase activity. (A) Baculovirus-expressed WT, K52R, P47A, and M299I BACH1 species were purified simultaneously and resolved on a 4–12% SDS-polyacrylamide gradient gel. Western blot analysis was performed by using a BACH1 specific polyclonal antibody. (B) Time course of ATPase activity of WT and BACH1 mutant species. (C) Helicase activity of WT and mutant BACH1 proteins (200 ng each) using a 19-mer-containing substrate. (D) Equivalent amounts of WT and M299I BACH1 were determined by quantitative Western blot analysis and protein measurements. Equal quantities of the two BACH1 proteins were incubated with a 99-mer-containing substrate. Results were quantitated with the assistance of a Molecular Dynamics STORM PhosphorImager and represented as percent released 32P-99-mer. Similar results were obtained in three independent experiments.
Similar articles
- BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function.
Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. Cantor SB, et al. Cell. 2001 Apr 6;105(1):149-60. doi: 10.1016/s0092-8674(01)00304-x. Cell. 2001. PMID: 11301010 - BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1.
Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmüller L. Dohrn L, et al. Biochem J. 2012 Feb 1;441(3):919-26. doi: 10.1042/BJ20110314. Biochem J. 2012. PMID: 22032289 - Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling.
Shiozaki EN, Gu L, Yan N, Shi Y. Shiozaki EN, et al. Mol Cell. 2004 May 7;14(3):405-12. doi: 10.1016/s1097-2765(04)00238-2. Mol Cell. 2004. PMID: 15125843 - Assessing the link between BACH1 and BRCA1 in the FA pathway.
Cantor SB, Andreassen PR. Cantor SB, et al. Cell Cycle. 2006 Jan;5(2):164-7. doi: 10.4161/cc.5.2.2338. Epub 2006 Jan 16. Cell Cycle. 2006. PMID: 16357529 Review. - Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair.
Cantor SB, Xie J. Cantor SB, et al. Environ Mol Mutagen. 2010 Jul;51(6):500-7. doi: 10.1002/em.20568. Environ Mol Mutagen. 2010. PMID: 20658644 Review.
Cited by
- FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response.
Xie J, Peng M, Guillemette S, Quan S, Maniatis S, Wu Y, Venkatesh A, Shaffer SA, Brosh RM Jr, Cantor SB. Xie J, et al. PLoS Genet. 2012 Jul;8(7):e1002786. doi: 10.1371/journal.pgen.1002786. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792074 Free PMC article. - BRIP1 (BACH1) variants and familial breast cancer risk: a case-control study.
Frank B, Hemminki K, Meindl A, Wappenschmidt B, Sutter C, Kiechle M, Bugert P, Schmutzler RK, Bartram CR, Burwinkel B. Frank B, et al. BMC Cancer. 2007 May 15;7:83. doi: 10.1186/1471-2407-7-83. BMC Cancer. 2007. PMID: 17504528 Free PMC article. - Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress.
Peng M, Cong K, Panzarino NJ, Nayak S, Calvo J, Deng B, Zhu LJ, Morocz M, Hegedus L, Haracska L, Cantor SB. Peng M, et al. Cell Rep. 2018 Sep 18;24(12):3251-3261. doi: 10.1016/j.celrep.2018.08.065. Cell Rep. 2018. PMID: 30232006 Free PMC article. - The Q motif of Fanconi anemia group J protein (FANCJ) DNA helicase regulates its dimerization, DNA binding, and DNA repair function.
Wu Y, Sommers JA, Loiland JA, Kitao H, Kuper J, Kisker C, Brosh RM Jr. Wu Y, et al. J Biol Chem. 2012 Jun 22;287(26):21699-716. doi: 10.1074/jbc.M112.351338. Epub 2012 May 10. J Biol Chem. 2012. PMID: 22582397 Free PMC article.
References
- Bork, P., Hofmann, K., Bucher, P., Neuwald, A. F., Altschul, S. F. & Koonin, E. V. (1997) FASEB J. 11, 68–76. - PubMed
- Scully, R., Ganesan, S., Vlasakova, K., Chen, J. J., Socolovsky, M. & Livingston, D. M. (1999) Mol. Cell 4, 1093–1099. - PubMed
- Snouwaert, J. N., Pace, A. J., Gowen, L. C., Xiao, A., Nichols, M. A., Xiong, Y. & Koller, B. H. (1999) FASEB J. 13, A1538–A1538.
- Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. (1999) Mol. Cell 4, 511–518. - PubMed
- Jasin, M. (2002) Oncogene 21, 8981–8993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous