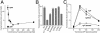Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage - PubMed (original) (raw)
Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage
Angela Ho et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Amyloid-beta precursor protein (APP), a type I membrane protein, is physiologically processed by alpha- or beta-secretases that cleave APP N-terminal to the transmembrane region. Extracellular alpha-/beta-cleavage of APP generates a large secreted N-terminal fragment, and a smaller cellular C-terminal fragment. Subsequent gamma-secretase cleavage in the transmembrane region of the C-terminal fragment induces secretion of small extracellular peptides, including Abeta40 and Abeta42, which are instrumental in the pathogenesis of Alzheimer's disease, and intracellular release of a cytoplasmic tail fragment. Although APP resembles a cell-surface receptor, no functionally active extracellular ligand for APP that might regulate its proteolytic processing has been described. We now show that F-spondin, a secreted signaling molecule implicated in neuronal development and repair, binds to the conserved central extracellular domain of APP and inhibits beta-secretase cleavage of APP. Our data indicate that F-spondin may be an endogenous regulator of APP cleavage, and suggest that the extracellular domains of APP are potential drug targets for interfering with beta-secretase cleavage.
Figures
Fig. 1.
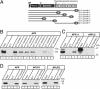
Binding of F-spondin to immobilized APP. (A) Domain structure of APP (Upper) and diagram of various APP vectors used for the present study (Lower). N-terminally, APP is composed of a signal peptide (SP), a CRD, a zinc-binding motif, acidic sequence regions, and a Kunitz domain. The center of APP is occupied by a large domain that contains no cysteine residues (referred to as CAPPD) and a short linker sequence that includes the cleavage sites for α- and β-secretases. C-terminally, APP contains a transmembrane region and a cytoplasmic tail. The constructs used here include Ig-fusion proteins of the entire extracellular region or CRD alone (Ig-APP.1 or Ig-APP.2 respectively), a GST-CAPPD fusion protein, and expression vectors that encode full-length APP, or APP in which the CRD or part of the CAPPD were deleted marked by dashed lines (pCMV-APPΔ1 or 2, respectively). Nonneuronal APP contains an alternatively spliced Kunitz domain that is absent from all APP constructs used here. (B) Affinity chromatography of secreted myc-tagged recombinant F-spondin pull-downs on immobilized APP proteins. Ig- and GST-fusion proteins of various fragments of APP, as indicated in A, were used to affinity-purify secreted myc-tagged F-spondin produced in the supernatant of transfected COS cells.
Fig. 2.
Binding of APP to immobilized F-spondin. (A) Domain structure of F-spondin (Upper) and constructs of F-spondin included in the various Ig-fusion vectors used for the present study (Lower). F-spondin is composed of an N-terminal signal peptide (SP), reelin-like and spondin domains, and six C-terminal thrombospondin repeats. The positions of the two N-glycosylation sites are indicated. (B) Pulldown of full-length APP695. APP695 was solubilized with 1% Triton X-100 from transfected COS cells and was bound to immobilized Ig-F spondin proteins containing full-length or parts of F-spondin (see A). (C) Pulldown of APP deletion mutants (see Fig. 1_A_ for extent of the deletions) with full-length Ig-F spondin. (D) Comparison of the ability of immobilized full-length F-spondin to affinity-purified APP, APLP1, and APLP2 expressed in transfected COS cells and visualized with antibodies to the C termini of indicated proteins.
Fig. 3.
Lack of an interaction of APP with Mindin. (A) Domain structure of F-spondin with Mindin. SP, signal peptide, a spondin-like domain; TSR, thrombospondin repeat. (B) Pulldown of myc-tagged Mindin with immobilized GST-CAPPD fusion protein. (C) Pulldown of APP with a Ig-Mindin fusion protein.
Fig. 4.
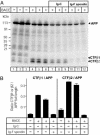
F-spondin inhibits cleavage of APP by BACE 1. (A) Immunoblot of HEK293 cells that were transfected without or with BACE 1, Ig-C, or Ig-F spondin as indicated. Experiments were carried out in triplicate to ensure reproducibility. Numbers on the left indicate positions of molecular mass markers. Note that BACE 1 cotransfection promotes production of two C-terminal fragments, termed CTFβ1 and CTFβ2. (B) Quantification of the results shown in A. Relative levels of full-length APP and of both CTFs were quantified using 125I-labeled secondary antibodies and PhosphorImager detection. Data shown are means ± SEM derived by dividing for each sample the signal for CTFβ1 or CTFβ2 by the APP signal.
Fig. 5.
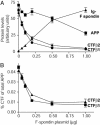
Titration of F-spondin-mediated inhibition of APP cleavage by BACE 1. (A) Relative levels of proteins expressed in an experiment similar to that described in Fig. 4. Increasing amounts of Ig-F spondin plasmid were cotransfected with constant amounts of APP and BACE 1. The levels of full-length APP and the CTFs of APP and of F-spondin were quantified by immunoblotting and are shown in arbitrary units. (B) Ratio of CTF to full-length APP as a function of increasing amount of F-spondin. CTF levels were corrected for APP expression. Data shown are means ± SEM from a representative experiment (n = 3) independently repeated multiple times.
Fig. 6.
Effect of F-spondin on APP-dependent transactivation of Gal4-Tip60-mediated transcription. (A) F-spondin inhibits APP-dependent transactivation. A constant amount of Gal4-Tip60, Fe65, and APP was cotransfected with increasing amounts of Ig-F spondin. Note that, without F-spondin, APP causes a strong stimulation of Gal4-Tip60-dependent transcription as described (7). F-spondin dramatically inhibits APP-dependent transactivation of transcription by Gal4-Tip60, such that even low concentrations of F-spondin (<100 ng of transfected plasmid) almost completely block the response. (B) Comparison of the effects of multiple Ig-fusion proteins on the APP-dependent transactivation of Gal4-Tip60. All proteins were expressed with 50 ng of cotransfected plasmids. (C) Increasing concentrations of APP are unable to rescue the F-spondin-dependent inhibition of APP-dependent transactivation of Gal4-Tip60. Constant amounts of Gal4-Tip60, Fe65, and Ig-C, Ig-N1β-1, or Ig-F spondin were cotransfected with increasing concentrations of APP. The bell-shaped dose–response curve under control conditions as reported (7) is probably due to dilution of transcription factors by increasing amounts of APP. Nevertheless, even at high concentrations of APP, F-spondin induces a relative inhibition of transactivation.
Similar articles
- Proteasome-mediated degradation of the C-terminus of the Alzheimer's disease beta-amyloid protein precursor: effect of C-terminal truncation on production of beta-amyloid protein.
Nunan J, Williamson NA, Hill AF, Sernee MF, Masters CL, Small DH. Nunan J, et al. J Neurosci Res. 2003 Nov 1;74(3):378-85. doi: 10.1002/jnr.10646. J Neurosci Res. 2003. PMID: 14598314 - F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein.
Hoe HS, Wessner D, Beffert U, Becker AG, Matsuoka Y, Rebeck GW. Hoe HS, et al. Mol Cell Biol. 2005 Nov;25(21):9259-68. doi: 10.1128/MCB.25.21.9259-9268.2005. Mol Cell Biol. 2005. PMID: 16227578 Free PMC article. - Functional interactions of APP with the apoE receptor family.
Hoe HS, Rebeck GW. Hoe HS, et al. J Neurochem. 2008 Sep;106(6):2263-71. doi: 10.1111/j.1471-4159.2008.05517.x. Epub 2008 Jun 28. J Neurochem. 2008. PMID: 18554321 Review. - Regulation of APP cleavage by alpha-, beta- and gamma-secretases.
Nunan J, Small DH. Nunan J, et al. FEBS Lett. 2000 Oct 13;483(1):6-10. doi: 10.1016/s0014-5793(00)02076-7. FEBS Lett. 2000. PMID: 11033346 Review.
Cited by
- Canonical and Non-canonical Reelin Signaling.
Bock HH, May P. Bock HH, et al. Front Cell Neurosci. 2016 Jun 30;10:166. doi: 10.3389/fncel.2016.00166. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27445693 Free PMC article. Review. - Proteolytic processing of Alzheimer's β-amyloid precursor protein.
Zhang H, Ma Q, Zhang YW, Xu H. Zhang H, et al. J Neurochem. 2012 Jan;120 Suppl 1(Suppl 1):9-21. doi: 10.1111/j.1471-4159.2011.07519.x. Epub 2011 Nov 28. J Neurochem. 2012. PMID: 22122372 Free PMC article. Review. - Interaction of the amyloid precursor protein-like protein 1 (APLP1) E2 domain with heparan sulfate involves two distinct binding modes.
Dahms SO, Mayer MC, Roeser D, Multhaup G, Than ME. Dahms SO, et al. Acta Crystallogr D Biol Crystallogr. 2015 Mar;71(Pt 3):494-504. doi: 10.1107/S1399004714027114. Epub 2015 Feb 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25760599 Free PMC article. - Amyloid precursor protein is an autonomous growth cone adhesion molecule engaged in contact guidance.
Sosa LJ, Bergman J, Estrada-Bernal A, Glorioso TJ, Kittelson JM, Pfenninger KH. Sosa LJ, et al. PLoS One. 2013 May 14;8(5):e64521. doi: 10.1371/journal.pone.0064521. Print 2013. PLoS One. 2013. PMID: 23691241 Free PMC article. - Regulated proteolysis of APP and ApoE receptors.
Hoe HS, Rebeck GW. Hoe HS, et al. Mol Neurobiol. 2008 Feb;37(1):64-72. doi: 10.1007/s12035-008-8017-0. Epub 2008 Apr 15. Mol Neurobiol. 2008. PMID: 18415033 Review.
References
- Price, D. L. & Sisodia, S. S. (1998) Annu. Rev. Neurosci. 21, 479-505. - PubMed
- Selkoe, D. J. (1998) Trends Cell Biol. 8, 447-453. - PubMed
- Bayer, T. A., Cappai, R., Masters, C. L., Beyreuther, K. & Multhaup, G. (1999) Mol. Psychiatry 4, 524-528. - PubMed
- Haass, C. & De Strooper, B. (1999) Science 286, 916-919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases