ARB: a software environment for sequence data - PubMed (original) (raw)
. 2004 Feb 25;32(4):1363-71.
doi: 10.1093/nar/gkh293. Print 2004.
Oliver Strunk, Ralf Westram, Lothar Richter, Harald Meier, Yadhukumar, Arno Buchner, Tina Lai, Susanne Steppi, Gangolf Jobb, Wolfram Förster, Igor Brettske, Stefan Gerber, Anton W Ginhart, Oliver Gross, Silke Grumann, Stefan Hermann, Ralf Jost, Andreas König, Thomas Liss, Ralph Lüssmann, Michael May, Björn Nonhoff, Boris Reichel, Robert Strehlow, Alexandros Stamatakis, Norbert Stuckmann, Alexander Vilbig, Michael Lenke, Thomas Ludwig, Arndt Bode, Karl-Heinz Schleifer
Affiliations
- PMID: 14985472
- PMCID: PMC390282
- DOI: 10.1093/nar/gkh293
ARB: a software environment for sequence data
Wolfgang Ludwig et al. Nucleic Acids Res. 2004.
Abstract
The ARB (from Latin arbor, tree) project was initiated almost 10 years ago. The ARB program package comprises a variety of directly interacting software tools for sequence database maintenance and analysis which are controlled by a common graphical user interface. Although it was initially designed for ribosomal RNA data, it can be used for any nucleic and amino acid sequence data as well. A central database contains processed (aligned) primary structure data. Any additional descriptive data can be stored in database fields assigned to the individual sequences or linked via local or worldwide networks. A phylogenetic tree visualized in the main window can be used for data access and visualization. The package comprises additional tools for data import and export, sequence alignment, primary and secondary structure editing, profile and filter calculation, phylogenetic analyses, specific hybridization probe design and evaluation and other components for data analysis. Currently, the package is used by numerous working groups worldwide.
Figures
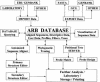
Figure 1
The interacting components and tools of the ARB software package and database.
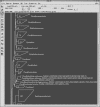
Figure 2
Example of a data visualization window. Bibliographic data stored in respective database fields are shown. The selection of database fields, extraction of data and the layout of the visualization window can be customized by the user.
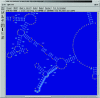
Figure 3
The ARB main window showing part of an ARB parsimony-generated dendrogram. The rectangles represent ‘online compressed’ monophyletic groups which can be ‘unfolded’ by mouse click. Database field entries such as taxonomic name, public database accession number and strain designation as reported in EMBL (1), RDP (3) and the European rRNA databases (DEW) (4,5) are visualized at the terminal nodes of the ‘unfolded’ Desulfohalobiaceae.
Figure 4
The ARB primary structure editor. As an example for highlighting a search string a probe target site is shown by background color. Perfect and mismatched pairing is color coded as well.
Figure 5
Secondary structure editor. The sequence selected in the primary structure editor (Fig. 4) is automatically fitted into a consensus secondary structure model.
Figure 6
Results of probe design and evaluation. Part of the primary structure alignment containing the probe target site is shown for the target organism Desulfohalobium retbaense and the non-target organisms containing the most similar sequence stretches.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases