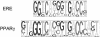Detection of functional DNA motifs via statistical over-representation - PubMed (original) (raw)
Comparative Study
. 2004 Feb 26;32(4):1372-81.
doi: 10.1093/nar/gkh299. Print 2004.
Affiliations
- PMID: 14988425
- PMCID: PMC390287
- DOI: 10.1093/nar/gkh299
Comparative Study
Detection of functional DNA motifs via statistical over-representation
Martin C Frith et al. Nucleic Acids Res. 2004.
Abstract
The interaction of proteins with DNA recognition motifs regulates a number of fundamental biological processes, including transcription. To understand these processes, we need to know which motifs are present in a sequence and which factors bind to them. We describe a method to screen a set of DNA sequences against a precompiled library of motifs, and assess which, if any, of the motifs are statistically over- or under-represented in the sequences. Over-represented motifs are good candidates for playing a functional role in the sequences, while under-representation hints that if the motif were present, it would have a harmful dysregulatory effect. We apply our method (implemented as a computer program called Clover) to dopamine-responsive promoters, sequences flanking binding sites for the transcription factor LSF, sequences that direct transcription in muscle and liver, and Drosophila segmentation enhancers. In each case Clover successfully detects motifs known to function in the sequences, and intriguing and testable hypotheses are made concerning additional motifs. Clover compares favorably with an ab initio motif discovery algorithm based on sequence alignment, when the motif library includes only a homolog of the factor that actually regulates the sequences. It also demonstrates superior performance over two contingency table based over-representation methods. In conclusion, Clover has the potential to greatly accelerate characterization of signals that regulate transcription.
Figures
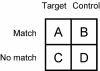
Figure 1
A 2×2 contingency table.
Figure 2
Pictogram representations of the ERE (3) and the Jaspar PPARγ motif (C Burge and F White,
http://genes.mit.edu/pictogram.html
).
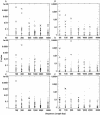
Figure 3
Detection by Clover of ERE motifs embedded in random DNA sequences of varying length. In all panels, the _P_-values of the 108 Jaspar motifs are plotted as dots. _P_-values of zero were increased to 0.001 to fit on the log scale. Crosses indicate the PPARγ motif, and circles indicate the six other ERE-like nuclear receptor motifs. (A) Results for 15 ERE-containing sequences with no decoy sequences. (B) Results for 15 ERE-containing sequences with five decoy sequences. (C) Results for 15 ERE-containing sequences with 15 decoy sequences.
Figure 4
Detection by contingency table based methods of EREs embedded in random DNA sequences of varying length. In all panels, the _P_-values of the 108 Jaspar motifs are plotted as dots. Crosses indicate the PPARγ motif, and circles indicate the six other ERE-like nuclear receptor motifs. (A, B, C) Motif counting method. Length 50 sequences were not analyzed because the number of possible locations is <1000 for some motifs, making the 0.1% threshold criterion impossible. (D, E, F) Sequence counting method. (A, D) Results for 15 ERE-containing sequences with no decoy sequences. (B, E) Results for 15 ERE-containing sequences with five decoy sequences. (C, F) Results for 15 ERE-containing sequences with 15 decoy sequences.
Similar articles
- Ab initio identification of putative human transcription factor binding sites by comparative genomics.
Corà D, Herrmann C, Dieterich C, Di Cunto F, Provero P, Caselle M. Corà D, et al. BMC Bioinformatics. 2005 May 2;6:110. doi: 10.1186/1471-2105-6-110. BMC Bioinformatics. 2005. PMID: 15865625 Free PMC article. - DNA motif representation with nucleotide dependency.
Chin F, Leung HC. Chin F, et al. IEEE/ACM Trans Comput Biol Bioinform. 2008 Jan-Mar;5(1):110-9. doi: 10.1109/TCBB.2007.70220. IEEE/ACM Trans Comput Biol Bioinform. 2008. PMID: 18245880 - Finding motifs from all sequences with and without binding sites.
Leung HC, Chin FY. Leung HC, et al. Bioinformatics. 2006 Sep 15;22(18):2217-23. doi: 10.1093/bioinformatics/btl371. Epub 2006 Jul 26. Bioinformatics. 2006. PMID: 16870937 - YMF: A program for discovery of novel transcription factor binding sites by statistical overrepresentation.
Sinha S, Tompa M. Sinha S, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3586-8. doi: 10.1093/nar/gkg618. Nucleic Acids Res. 2003. PMID: 12824371 Free PMC article. - DNA binding sites: representation and discovery.
Stormo GD. Stormo GD. Bioinformatics. 2000 Jan;16(1):16-23. doi: 10.1093/bioinformatics/16.1.16. Bioinformatics. 2000. PMID: 10812473 Review.
Cited by
- Nuclear RNA sequencing of the mouse erythroid cell transcriptome.
Mitchell JA, Clay I, Umlauf D, Chen CY, Moir CA, Eskiw CH, Schoenfelder S, Chakalova L, Nagano T, Fraser P. Mitchell JA, et al. PLoS One. 2012;7(11):e49274. doi: 10.1371/journal.pone.0049274. Epub 2012 Nov 29. PLoS One. 2012. PMID: 23209567 Free PMC article. - Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function.
Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Sakabe NJ, et al. Hum Mol Genet. 2012 May 15;21(10):2194-204. doi: 10.1093/hmg/dds034. Epub 2012 Feb 10. Hum Mol Genet. 2012. PMID: 22328084 Free PMC article. - Measuring similarities between transcription factor binding sites.
Kielbasa SM, Gonze D, Herzel H. Kielbasa SM, et al. BMC Bioinformatics. 2005 Sep 28;6:237. doi: 10.1186/1471-2105-6-237. BMC Bioinformatics. 2005. PMID: 16191190 Free PMC article. - A survey of motif discovery methods in an integrated framework.
Sandve GK, Drabløs F. Sandve GK, et al. Biol Direct. 2006 Apr 6;1:11. doi: 10.1186/1745-6150-1-11. Biol Direct. 2006. PMID: 16600018 Free PMC article. - Set cover-based methods for motif selection.
Li Y, Liu Y, Juedes D, Drews F, Bunescu R, Welch L. Li Y, et al. Bioinformatics. 2020 Feb 15;36(4):1044-1051. doi: 10.1093/bioinformatics/btz697. Bioinformatics. 2020. PMID: 31665223 Free PMC article.
References
- Stormo G.D. (2000). DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. - PubMed
- Pennacchio L.A. and Rubin,E.M. (2001). Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet., 2, 100–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-CA81157/CA/NCI NIH HHS/United States
- 1R01HG03110-01/HG/NHGRI NIH HHS/United States
- P20 GM066401/GM/NIGMS NIH HHS/United States
- R01 CA081157/CA/NCI NIH HHS/United States
- R01 CA081157-05/CA/NCI NIH HHS/United States
- NS37403/NS/NINDS NIH HHS/United States
- R01 HG003110/HG/NHGRI NIH HHS/United States
- 1P20GM066401-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases