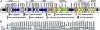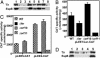Dissecting virulence: systematic and functional analyses of a pathogenicity island - PubMed (original) (raw)
. 2004 Mar 9;101(10):3597-602.
doi: 10.1073/pnas.0400326101. Epub 2004 Feb 26.
José L Puente, Samantha Gruenheid, Yuling Li, Bruce A Vallance, Alejandra Vázquez, Jeannette Barba, J Antonio Ibarra, Paul O'Donnell, Pavel Metalnikov, Keith Ashman, Sansan Lee, David Goode, Tony Pawson, B Brett Finlay
Affiliations
- PMID: 14988506
- PMCID: PMC373508
- DOI: 10.1073/pnas.0400326101
Dissecting virulence: systematic and functional analyses of a pathogenicity island
Wanyin Deng et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Bacterial pathogenicity islands (PAI) often encode both effector molecules responsible for disease and secretion systems that deliver these effectors to host cells. Human enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli, and the mouse pathogen Citrobacter rodentium (CR) possess the locus of enterocyte effacement (LEE) PAI. We systematically mutagenized all 41 CR LEE genes and functionally characterized these mutants in vitro and in a murine infection model. We identified 33 virulence factors, including two virulence regulators and a hierarchical switch for type III secretion. In addition, 7 potential type III effectors encoded outside the LEE were identified by using a proteomics approach. These non-LEE effectors are encoded by three uncharacterized PAIs in EHEC O157, suggesting that these PAIs act cooperatively with the LEE in pathogenesis. Our findings provide significant insights into bacterial virulence mechanisms and disease.
Figures
Fig. 1.
Both Ler and Orf11 are required for expression of LEE genes in CR. (A) Genetic organization of CR LEE (7). (B) Expression of Tir and EspB in WT CR and its 41 LEE mutants. Whole-cell lysates of bacteria grown in DMEM were analyzed by 10% SDS/PAGE and Western blot with anti-Tir and anti-EspB sera.
Fig. 2.
Orf11 and Orf10 regulate ler expression in CR. (A) Western blot with anti-Tir and anti-EspB sera of total lysates of bacteria grown in DMEM. Lane 1, WT CR; lane 2, Δ_orf11_; lane 3, Δ_ler_Δ_orf11._ Also shown are CR Δ_orf11_ complemented by orf11 from CR (pCRorf11, lane 4), EHEC (pEHorf11, lane 5), or EPEC (pEPorf11, lane 6); and CR Δ_ler_Δ_orf11_ double mutant complemented by CR ler (lane 7) or orf11 (lane 8). (B) Orf11 positively regulates ler expression. The transcriptional activity directed by the ler-cat fusion in pLEE1/Ler-CAT was determined in CR WT, Δ_ler_, Δ_orf10_, and Δ_orf11_ grown in DMEM for 6 h. The data are the average of three experiments. (C) Orf11 positively regulates the expression of LEE2 and LEE5 operons by activating ler expression. The activity directed by LEE2 (pLEE2-CAT) and LEE5 (pLEE5-CAT) transcriptional fusions was measured in CR WT, Δ_ler_, Δ_orf10_, and Δ_orf11_ as described above. (D) Orf10 acts as a negative regulator of LEE gene expression when expressed from a plasmid. Whole-cell lysates of WT CR carrying pCR2.1-TOPO (the cloning vector, lane 1), pCRorf10-2HA (2HA-tagged orf10, lane 2), pCRorf10 (CR orf10 with its own promoter, lane 3), pCRorf10Plac (P_lac_-driven CR orf10, lane 4), and pCRorf10orf11 (CR orf10 and orf11 with their own promoter, lane 5) were analyzed as for A.
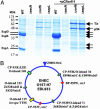
Fig. 3.
Type III secretion by WT CR and its 41 LEE mutants. (A) General protein secretion profile of CR and its mutants. (B) Tir and EspB secretion analyzed by Western blot with anti-Tir and anti-EspB sera. (C) Secretion profile of Δ_sepL_, Δ_rorf6_, Δ_escN_ (TTS mutant), and their double mutants. Secreted proteins were concentrated from supernatants of bacterial cultures grown in DMEM and analyzed by 12% SDS/PAGE and Coomassie blue G250 staining (A and C) or Western blot (B).
Fig. 4.
Identification of both LEE- and non-LEE-encoded proteins secreted by the LEE-encoded TTSS. (A) Effect of overexpressing CR orf11 on TTS in WT CR and its Δ_sepL_ or Δ_rorf6_ mutants. Secreted proteins were analyzed by 15% SDS/PAGE and Coomassie blue staining. The additional type III secreted proteins by Δ_sepL_ and Δ_rorf6_ carrying pCRorf11 are indicated by arrows and were characterized by proteomic analyses (Table 2 and Fig. 8). (B) A diagram showing locations of the O-islands encoding the six identified non-LEE effectors in the EHEC O157:H7 genome (3). Also shown are the locations of the Shiga toxin genes (stx), the LEE, the _inv-spa_-like TTSS, and the associated prophages (CP- and BP-933).
Similar articles
- Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium.
Kelly M, Hart E, Mundy R, Marchès O, Wiles S, Badea L, Luck S, Tauschek M, Frankel G, Robins-Browne RM, Hartland EL. Kelly M, et al. Infect Immun. 2006 Apr;74(4):2328-37. doi: 10.1128/IAI.74.4.2328-2337.2006. Infect Immun. 2006. PMID: 16552063 Free PMC article. - Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7.
Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. Gruenheid S, et al. Mol Microbiol. 2004 Mar;51(5):1233-49. doi: 10.1046/j.1365-2958.2003.03911.x. Mol Microbiol. 2004. PMID: 14982621 - Bacteriophage Transcription Factor Cro Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli.
Hernandez-Doria JD, Sperandio V. Hernandez-Doria JD, et al. Cell Host Microbe. 2018 May 9;23(5):607-617.e6. doi: 10.1016/j.chom.2018.04.007. Cell Host Microbe. 2018. PMID: 29746832 Free PMC article. - [Plasticity of bacterial genomes: pathogenicity islands and the locus of enterocyte effacement (LEE)].
Kirsch P, Jores J, Wieler LH. Kirsch P, et al. Berl Munch Tierarztl Wochenschr. 2004 Mar-Apr;117(3-4):116-29. Berl Munch Tierarztl Wochenschr. 2004. PMID: 15046458 Review. German. - The Tip of the Iceberg: On the Roles of Regulatory Small RNAs in the Virulence of Enterohemorrhagic and Enteropathogenic Escherichia coli.
Bhatt S, Egan M, Jenkins V, Muche S, El-Fenej J. Bhatt S, et al. Front Cell Infect Microbiol. 2016 Sep 21;6:105. doi: 10.3389/fcimb.2016.00105. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27709103 Free PMC article. Review.
Cited by
- Involvement of PatE, a prophage-encoded AraC-like regulator, in the transcriptional activation of acid resistance pathways of enterohemorrhagic Escherichia coli strain EDL933.
Bender JK, Praszkier J, Wakefield MJ, Holt K, Tauschek M, Robins-Browne RM, Yang J. Bender JK, et al. Appl Environ Microbiol. 2012 Aug;78(15):5083-92. doi: 10.1128/AEM.00617-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582067 Free PMC article. - In vitro and in vivo model systems for studying enteropathogenic Escherichia coli infections.
Law RJ, Gur-Arie L, Rosenshine I, Finlay BB. Law RJ, et al. Cold Spring Harb Perspect Med. 2013 Mar 1;3(3):a009977. doi: 10.1101/cshperspect.a009977. Cold Spring Harb Perspect Med. 2013. PMID: 23457294 Free PMC article. Review. - Enteropathogenic Escherichia coli type III effectors EspG and EspG2 alter epithelial paracellular permeability.
Matsuzawa T, Kuwae A, Abe A. Matsuzawa T, et al. Infect Immun. 2005 Oct;73(10):6283-9. doi: 10.1128/IAI.73.10.6283-6289.2005. Infect Immun. 2005. PMID: 16177299 Free PMC article. - Efficient translocation of EspC into epithelial cells depends on enteropathogenic Escherichia coli and host cell contact.
Vidal JE, Navarro-García F. Vidal JE, et al. Infect Immun. 2006 Apr;74(4):2293-303. doi: 10.1128/IAI.74.4.2293-2303.2006. Infect Immun. 2006. PMID: 16552060 Free PMC article. - Bacterial-fungal interactions and their impact on microbial pathogenesis.
MacAlpine J, Robbins N, Cowen LE. MacAlpine J, et al. Mol Ecol. 2023 May;32(10):2565-2581. doi: 10.1111/mec.16411. Epub 2022 Mar 14. Mol Ecol. 2023. PMID: 35231147 Free PMC article. Review.
References
- Frankel, G., Phillips, A. D., Rosenshine, I., Dougan, G., Kaper, J. B. & Knutton, S. (1998) Mol. Microbiol. 30, 911-921. - PubMed
- Perna, N. T., Plunkett, G., 3rd, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. - PubMed
- Hacker, J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54, 641-679. - PubMed
- Elliott, S. J., Wainwright, L. A., McDaniel, T. K., Jarvis, K. G., Deng, Y. K., Lai, L. C., McNamara, B. P., Donnenberg, M. S. & Kaper, J. B. (1998) Mol. Microbiol. 28, 1-4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases