A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea - PubMed (original) (raw)
Figure 1
Genomic organization of the herA, mre11, rad50 and nurA genes from thermophilic archaea. Sulfolobales designed S.acidocaldarius, S.tokodaii and S.solfataricus P2, and Pyrococcales designed P.abyssi, P.horikoshii OT3 and P.furiosus.
Figure 2
Multiple alignment of the HerA protein family with selected proteins of the FtsK, TrwB/VirB4 and VirD4 ATPase families. Highly conserved residues are shown with bold type. The positions of the first and last residues of the aligned segments in the respective proteins are indicated by numbers. Poorly conserved regions not included in the alignment are denoted by the number of amino acid residues in parentheses. The secondary structure assignments are from the crystal structure of TrwB (PDB code 1E9R): H indicates α-helix and E indicates extended conformation (β-strand). The sequences are arranged in the following groups (designated to the right of the alignment): I, HerA proteins encoded within conserved archaeal operons; II, the remaining archaeal HerA proteins; III, bacterial HerA proteins; IV, bacteriophage ATPase; V, FtsK family ATPases; VI, TrwB/VirB4 family ATPases; VII, VirD4 family ATPases. The HerA family is represented in its entirety; for the other families, only selected sequences are shown. The sequences are denoted with the gene name followed by abbreviated species name, and the GI numbers. The S.acidocaldarius sequence has been submitted to the DDBJ/EMBL/GenBank databases under accession no. CAE51870. Species abbreviations are as follows: Archaea: Af, Archaeoglobus fulgidus; Ape, Aeropyrum pernix; Hsp, Halobacterium sp.; Mac, Methanosarcina acetivorans; Mj, Methanocaldococcus jannaschii; Mka, Methanopyrus kandleri; Mma, Methanosarcina mazei; Mta, Methanothermobacter thermoautotrophicus; Pab, Pyrococcus abyssi; Paer, Pyrobaculum aerophilum; Sac, Sulfolobus acidocaldarius; Sso, Sulfolobus solfataricus; Tac, Thermoplasma acidophilum; Tvo, Thermoplasma volcanium. Bacteria: Aae, Aquifex aeolicus; Atu, Agrobacterium tumefaciens; Bjap, Bradirhizobium japonicum; Bme, Brucella melitensis; Bper, Bordetella pertussis; Cau, Chloroflexus aurantiacus; Cje, Campylobacter jejuni; Ct, Chlamydia trachomatis; Ec, Escherichia coli; Fnu, Fusobacterium nucleatum; Hp, Helicobacter pylori; Lpne, Legionella pneumophila; Mtu, Mycobacterium tuberculosis; Npu, Nostoc punctiforme; Pae, Pseudomonas aeruginosa; Pput, Pseudomonas putida; Rme, Ralstonia metallidurans; Rsph, Rhodobacter sphaeroides; Rp, Rickettsia prowazekii; Sau, Staphylococcus aureus; Sme, Sinorhizobium meliloti; Ssp, Synechocystis sp.; Sty, Salmonella typhi; Tel, Thermosynechococcus elongatus BP-1; Tp, Treponema pallidum; Wol, Wolinella succinogenes.
Figure 2
Multiple alignment of the HerA protein family with selected proteins of the FtsK, TrwB/VirB4 and VirD4 ATPase families. Highly conserved residues are shown with bold type. The positions of the first and last residues of the aligned segments in the respective proteins are indicated by numbers. Poorly conserved regions not included in the alignment are denoted by the number of amino acid residues in parentheses. The secondary structure assignments are from the crystal structure of TrwB (PDB code 1E9R): H indicates α-helix and E indicates extended conformation (β-strand). The sequences are arranged in the following groups (designated to the right of the alignment): I, HerA proteins encoded within conserved archaeal operons; II, the remaining archaeal HerA proteins; III, bacterial HerA proteins; IV, bacteriophage ATPase; V, FtsK family ATPases; VI, TrwB/VirB4 family ATPases; VII, VirD4 family ATPases. The HerA family is represented in its entirety; for the other families, only selected sequences are shown. The sequences are denoted with the gene name followed by abbreviated species name, and the GI numbers. The S.acidocaldarius sequence has been submitted to the DDBJ/EMBL/GenBank databases under accession no. CAE51870. Species abbreviations are as follows: Archaea: Af, Archaeoglobus fulgidus; Ape, Aeropyrum pernix; Hsp, Halobacterium sp.; Mac, Methanosarcina acetivorans; Mj, Methanocaldococcus jannaschii; Mka, Methanopyrus kandleri; Mma, Methanosarcina mazei; Mta, Methanothermobacter thermoautotrophicus; Pab, Pyrococcus abyssi; Paer, Pyrobaculum aerophilum; Sac, Sulfolobus acidocaldarius; Sso, Sulfolobus solfataricus; Tac, Thermoplasma acidophilum; Tvo, Thermoplasma volcanium. Bacteria: Aae, Aquifex aeolicus; Atu, Agrobacterium tumefaciens; Bjap, Bradirhizobium japonicum; Bme, Brucella melitensis; Bper, Bordetella pertussis; Cau, Chloroflexus aurantiacus; Cje, Campylobacter jejuni; Ct, Chlamydia trachomatis; Ec, Escherichia coli; Fnu, Fusobacterium nucleatum; Hp, Helicobacter pylori; Lpne, Legionella pneumophila; Mtu, Mycobacterium tuberculosis; Npu, Nostoc punctiforme; Pae, Pseudomonas aeruginosa; Pput, Pseudomonas putida; Rme, Ralstonia metallidurans; Rsph, Rhodobacter sphaeroides; Rp, Rickettsia prowazekii; Sau, Staphylococcus aureus; Sme, Sinorhizobium meliloti; Ssp, Synechocystis sp.; Sty, Salmonella typhi; Tel, Thermosynechococcus elongatus BP-1; Tp, Treponema pallidum; Wol, Wolinella succinogenes.
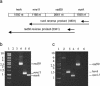
Figure 3
Co-transcription of S.acidocaldarius herA, mre11, rad50 and nurA genes. (a) Schematic representation of RT–PCR products obtained using either a nurA_- or a rad50_-specific primer for the RT reaction. (b and c) Analyses of RT–PCR products. (b) Lane 1, DNA ladder; lane 2, control PCR using 5′_mre11 and 3′_nurA primers on S.acidocaldarius total RNA; lanes 3–6, PCR products obtained with nurA reverse products and the following primers: lane 3, 5′mre11 and 3′nurA primers; lane 4, rad50 primers; lane 5, mre11 primers; lane 6, nurA primers. (c) Lane 1, DNA ladder; lanes 2–5, PCR products obtained with rad50 reverse products and the following primers: lane 2, 5′herA and 3′rad50 primers; lane 3, herA primers; lane 4, mre11 primers; lane 5, rad50 primers; lane 6, control PCR using 5′herA and 3′rad50 primers on S.acidocaldarius total RNA.
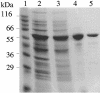
Figure 4
Purification of recombinant HerA protein. Purification steps were analyzed by SDS–PAGE and Coomassie blue staining. Lane 1, molecular weight standards; lane 2, overproducing extracts; lane 3, soluble fraction of overproducing extracts; lane 4, Ni2+-NTA agarose pool; lane 5, Source 30S pool.
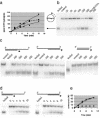
Figure 5
ATPase and helicase activities associated with HerA protein. (a) Time course of HerA ATPase activity: ATP hydrolysis was followed at 70°C for the indicated times in the absence of DNA (triangles) or in the presence of single-stranded DNA (squares) or double-stranded DNA (circles). (b) Helicase activity recovered with a primed circular single-stranded DNA substrate and 20–300 nM wild-type HerA protein or 300 nM HerAK153A protein in 30 min at 70°C. (c) Helicase activity recovered after a 30 min incubation at 70°C using 5′ overhang, 3′ overhang or blunt linear DNA substrates in the presence of 100–500 nM HerA protein. (d and e) Time course of HerA helicase activity. HerA (400 nM) was incubated at 70°C with 5′ overhang (triangles) or 3′ overhang (circles) DNA substrates for the time points shown. Squares correspond to the quantification of the control experiment [data not shown in (d)] performed without enzyme. In (b) and (c), control lanes correspond to the DNA substrate incubated for 30 min at 70°C without enzyme and in (b), (c) and (d), boiled lanes correspond to the heat-denatured substrate.