Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export - PubMed (original) (raw)
Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export
Rafael Bernad et al. Mol Cell Biol. 2004 Mar.
Abstract
Nuclear pore complexes (NPCs) traverse the nuclear envelope (NE), providing a channel through which nucleocytoplasmic transport occurs. Nup358/RanBP2, Nup214/CAN, and Nup88 are components of the cytoplasmic face of the NPC. Here we show that Nup88 localizes midway between Nup358 and Nup214 and physically interacts with them. RNA interference of either Nup88 or Nup214 in human cells caused a strong reduction of Nup358 at the NE. Nup88 and Nup214 showed an interdependence at the NPC and were not affected by the absence of Nup358. These data indicate that Nup88 and Nup214 mediate the attachment of Nup358 to the NPC. We show that localization of the export receptor CRM1 at the cytoplasmic face of the NE is Nup358 dependent and represents its empty state. Also, removal of Nup358 causes a distinct reduction in nuclear export signal-dependent nuclear export. We propose that Nup358 provides both a platform for rapid disassembly of CRM1 export complexes and a binding site for empty CRM1 recycling into the nucleus.
Figures
FIG. 1.
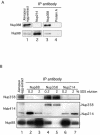
Xenopus Nup88 is encoded by two genes and is phosphorylated, and it localizes adjacent to Nup214/CAN and Nup358/RanBP2. (A) Western blot of Xenopus egg extracts probed with anti-XNup88. The asterisk represents a nonspecific cross-reacting band. (B) Immunofluorescence of Xenopus A6 cells probed with anti-XNup88 to show specific staining of the NE. (C) A phylogram showing divergence of the two genes encoding Nup88 in X. laevis and their conservation with respect to human Nup88 and rat Nup84. (D) Western blot of Xenopus egg extracts before (−) and after (+) treatment with lambda protein phosphatase (lambda-PPase) probed with anti-XNup88. (E) Representative scanning EM of an isolated Xenopus oocyte NE labeled with anti-XNup88, which was secondarily labeled with 10-nm colloidal gold. Bar = 50 nm. (F) Representative TEM of a 70-nm cross-section through an isolated Xenopus oocyte NE labeled with anti-XNup88 and secondarily labeled with 10-nm colloidal gold. N, nucleus; C, cytoplasm. Bar = 50 nm. (G) Summary diagram of the NPC displaying the mean localization of Nup88. Bar = 50 nm. Error bars represent standard deviations of the mean.
FIG. 2.
(A) Nup88 is coimmunoprecipitated with both Nup214/CAN and Nup358/RanBP2. Antibodies to Nup214, Nup358, or protein A-Sepharose were incubated with Xenopus egg extract, and coimmunoprecipitating proteins analyzed by labeling a Western blot with anti-Nup358 or anti-XNup88. (B) Antibodies to Nup88 (lanes 2 and 3), Nup358 (lanes 4 and 5), and Nup214 (lanes 6 and 7) were used to coimmunoprecipitate protein complexes from Xenopus egg extracts. Proteins were isolated using protein A-Sepharose beads, and coimmunoprecipitating proteins were eluted using 0.2% (lanes 2, 4, and 6) followed by 2% (lanes 3, 5, and 7) SDS and analyzed by labeling a Western blot with anti-Nup358 (upper panel), MAb 414 (middle panel), and anti-Nup88 (lower panel).
FIG.3.
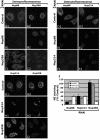
Knockdown of Nup88 or Nup214/CAN causes a decrease in Nup358/RanBP2 at the NE. Immunofluorescence of HeLa cells after knockdown of Nup88 (B and E), Nup214 (F and H), and Nup358 (C and I) is shown. Cells were fluorescently double labeled with anti-hNup88 and anti-hNup358F (A, B, and C), anti-hNup88 and anti-CAN9977 (D, E, and F), or anti-CAN9977 and anti-Nup358V antibodies (G, H, and I). The A, D, and G panels show control levels 72 h after transfection with empty pSUPER vector. (J) Graphic representation of the results to show fluorescence levels of Nup88, Nup214, and Nup358 after knockdown of each individual nucleoporin as a percentage of the negative control. Nup358 analysis was performed using two different antibodies, leading to similar results.
FIG. 4.
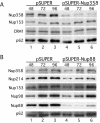
Efficient knockdown of Nup88 and Nup358/RanBP2 using RNAi and coreduction of Nup214/CAN on knockdown of Nup88. (A) Western blot of MCF-7 cells transfected by electroporation with pSUPER-Nup358 compared to the pSUPER negative control collected 48, 72, and 96 h posttransfection. The blot was probed with anti-Nup358V, MAb 414, and anti-CRM1. (B) Western blot of HeLa cells transfected by Fugene with pSUPER-Nup88 compared to the pSUPER negative control collected 48, 72, and 96 h posttransfection. The blot was probed with anti-Nup358V, anti-hNup88, anti-hNup214, anti-Nup98, and MAb 414.
FIG. 5.
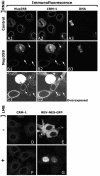
CRM1 is mislocalized from the cytoplasmic side of the NE in Nup358 knocked-down cells but not when cargo substrate binding is inhibited. HeLa cells were fixed and permeabilized with 0.001% digitonin 72 h posttranfection with pSUPER (A) or pSUPER-Nup358 (B and C). Antibodies for Nup358, CRM1, and DNA were applied. (B and C) Nup358 knocked-down cells, indicated by arrows, show reduced CRM1 NE staining at the accessible side of the NE. Under these conditions, DNA antibodies can only access dividing cells (A3 and B3). CRM1 localization at the cytoplasmic side of the NE was not altered upon leptomycin B (LMB) treatment of MCF-7 cells (D and F). Treatment was sufficient to abolish CRM1-mediated export of a REV-GFP construct (E and G).
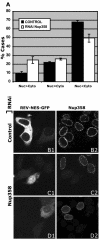
FIG. 6.
Nup358/RanBP2 knocked-down cells are less efficient in CRM1-mediated export of Rev-GFP. (A) Rev-GFP-transfected MCF-7 cells were scored for predominant accumulation of GFP in the cytoplasm (Nuc
<cyto), the="" nucleus="" (nuc="">Cyto), or equal distribution (Nuc=Cyto) between the two compartments under control (black bars) or Nup358 RNAi (white bars) conditions. The mean distribution in three independent experiments is shown; error bars represent standard errors. Illustrative images of Nup358 show a strong (C1 and C2) and weak (D1 and D2) export defect. Empty pSUPER-transfected cells are shown in panels B1 and B2.</cyto),>
Similar articles
- Nup214 is required for CRM1-dependent nuclear protein export in vivo.
Hutten S, Kehlenbach RH. Hutten S, et al. Mol Cell Biol. 2006 Sep;26(18):6772-85. doi: 10.1128/MCB.00342-06. Mol Cell Biol. 2006. PMID: 16943420 Free PMC article. - Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export.
Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. Bernad R, et al. J Biol Chem. 2006 Jul 14;281(28):19378-86. doi: 10.1074/jbc.M512585200. Epub 2006 May 4. J Biol Chem. 2006. PMID: 16675447 - The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila.
Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. Xylourgidis N, et al. J Cell Sci. 2006 Nov 1;119(Pt 21):4409-19. doi: 10.1242/jcs.03201. Epub 2006 Oct 10. J Cell Sci. 2006. PMID: 17032737 - Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease.
Jamali T, Jamali Y, Mehrbod M, Mofrad MR. Jamali T, et al. Int Rev Cell Mol Biol. 2011;287:233-86. doi: 10.1016/B978-0-12-386043-9.00006-2. Int Rev Cell Mol Biol. 2011. PMID: 21414590 Review. - [Structural basis for assembly and disassembly of the CRM1 nuclear export complex and its application to drug development].
Koyama M, Matsuura Y. Koyama M, et al. Seikagaku. 2015 Feb;87(1):41-8. Seikagaku. 2015. PMID: 26571554 Review. Japanese. No abstract available.
Cited by
- Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold.
Gaik M, Flemming D, von Appen A, Kastritis P, Mücke N, Fischer J, Stelter P, Ori A, Bui KH, Baßler J, Barbar E, Beck M, Hurt E. Gaik M, et al. J Cell Biol. 2015 Feb 2;208(3):283-97. doi: 10.1083/jcb.201411003. J Cell Biol. 2015. PMID: 25646085 Free PMC article. - Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins.
Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR. Takeda A, et al. J Biol Chem. 2010 May 21;285(21):16248-57. doi: 10.1074/jbc.M109.048785. Epub 2010 Mar 16. J Biol Chem. 2010. PMID: 20233715 Free PMC article. - Characterization of the role of the tumor marker Nup88 in mitosis.
Hashizume C, Nakano H, Yoshida K, Wong RW. Hashizume C, et al. Mol Cancer. 2010 May 24;9:119. doi: 10.1186/1476-4598-9-119. Mol Cancer. 2010. PMID: 20497554 Free PMC article. - Pathogenic Variants in NUP214 Cause "Plugged" Nuclear Pore Channels and Acute Febrile Encephalopathy.
Fichtman B, Harel T, Biran N, Zagairy F, Applegate CD, Salzberg Y, Gilboa T, Salah S, Shaag A, Simanovsky N, Ayoubieh H, Sobreira N, Punzi G, Pierri CL, Hamosh A, Elpeleg O, Harel A, Edvardson S. Fichtman B, et al. Am J Hum Genet. 2019 Jul 3;105(1):48-64. doi: 10.1016/j.ajhg.2019.05.003. Epub 2019 Jun 6. Am J Hum Genet. 2019. PMID: 31178128 Free PMC article. - Nuclear export of the pre-60S ribosomal subunit through single nuclear pores observed in real time.
Ruland JA, Krüger AM, Dörner K, Bhatia R, Wirths S, Poetes D, Kutay U, Siebrasse JP, Kubitscheck U. Ruland JA, et al. Nat Commun. 2021 Oct 27;12(1):6211. doi: 10.1038/s41467-021-26323-7. Nat Commun. 2021. PMID: 34707094 Free PMC article.
References
- Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55-66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous