Structural characterization of the SARS-coronavirus spike S fusion protein core - PubMed (original) (raw)
Structural characterization of the SARS-coronavirus spike S fusion protein core
Brian Tripet et al. J Biol Chem. 2004.
Abstract
The spike (S) glycoprotein of coronaviruses mediates viral entry into host cells. It is a type 1 viral fusion protein that characteristically contains two heptad repeat regions, denoted HR-N and HR-C, that form coiled-coil structures within the ectodomain of the protein. Previous studies have shown that the two heptad repeat regions can undergo a conformational change from their native state to a 6-helix bundle (trimer of dimers), which mediates fusion of viral and host cell membranes. Here we describe the biophysical analysis of the two predicted heptad repeat regions within the severe acute respiratory syndrome coronavirus S protein. Our results show that in isolation the HR-N region forms a stable alpha-helical coiled coil that associates in a tetrameric state. The HR-C region in isolation formed a weakly stable trimeric coiled coil. When mixed together, the two peptide regions (HR-N and HR-C) associated to form a very stable alpha-helical 6-stranded structure (trimer of heterodimers). Systematic peptide mapping showed that the site of interaction between the HR-N and HR-C regions is between residues 916-950 of HR-N and residues 1151-1185 of HR-C. Additionally, interchain disulfide bridge experiments showed that the relative orientation of the HR-N and HR-C helices in the complex was antiparallel. Overall, the structure of the hetero-stranded complex is consistent with the structures observed for other type 1 viral fusion proteins in their fusion-competent state.
Figures
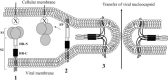
Fig. 1
Schematic representation of the three different states of the coronavirus S fusion protein during viral entry.State 1 (native), the S protein is denoted S1 and S2 for its N- and C-terminal domains; state 2 (intermediate state), the N-terminal (S1) domain is dissociated (shedding) to expose the fusion peptide (FP) region, and state 3 (collapsed 6-helix bundle or fusion active state), the collapsed S2 domain draws the viral and cellular membranes together causing fusion and release of the viral nucleocapsid into the host cell. HR-N and HR-C denote coiled coils at the N terminus and C terminus of the S2 domain. X denotes the host cell surface receptor.
Fig. 2
Coiled-coil prediction analysis of the SARS-CoV S glycoprotein.A, plot of the predicted stability versus sequence position for the SARS-CoV S protein using the coiled-coil prediction algorithm STABLECOIL (53) with a 35-residue window width. The two regions of interest to this study are denoted HR-N (for N-terminal heptad repeat) and HR-C (for C-terminal heptad repeat). B, schematic representation of the relative locations of the predicted coiled-coils HR-N and HR-C within the S protein of the SARS-CoV. The location of the predicted transmembrane spanning region and fusion peptide region are also shown. HR-N can be divided into three sub-regions (regions 1–3) based on the assignment of the a and d positions. Changes in the a and d register are denoted as a frameshift/hinge region.
Fig. 3
Top, sequence of the predicted HR-N region from residues 882–1011 of the SARS-CoV S protein. The a and d positions of the strongest predicted coiled-coil heptad repeats (abcdefg)n are shown above the sequence. The a and d positions of the alternate weaker scoring heptad repeat is shown below the sequence. Middle, the names and sequence regions (denoted in parentheses) of the HR-N peptides used in this study. The rectangle denotes the HR-N sequence from residues 882 to 1011. The hatched areas denote regions 1–3 (Fig. 2). The bars below the HR-N domain indicate the locations of the peptides within the HR-N region used in this study. Bottom, the sequence of the predicted HR-C domain from residues 1147–1185. The a and d positions are shown above the sequence. The names, sequences, and locations (bars) of the HR-C peptides used in this study are shown below the HR-C domain.
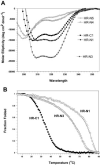
Fig. 4
A, CD spectra of peptides corresponding to the HR-N and HR-C of the SARS-CoV S protein. Spectra were recorded in a 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer. Peptide concentrations were 100 μ
m
for HR-N3 and HR-C1, and 15 μ
m
for HR-N1, HR-N4, and HR-N5. B, temperature denaturation profiles of the HR-N and HR-C peptides monitored by CD at 222 nm in a 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer. Concentrations were 15 μ
m
for HR-N1 and 100 μ
m
for HR-N3 and HR-C1 peptides. The fraction folded (ff) of each peptide was calculated as ff = ([θ] – [θ]u)/([θ]n – [θ]u), where [θ] is the observed mean residue ellipticity at 222 nm at any particular temperature, and [θ]n and [θ]u are the mean residue ellipticities at 222 nm of the native folded state at 4 °C and unfolded states, respectively.
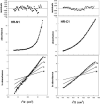
Fig. 5
Equilibrium ultracentrifugation analysis of the HR-N and HR-C peptides.Left side, middle, a plot absorbance versus radial distance squared divided by 2 for HR-N1 at 22 °C and 26,000 rpm in a 0.1
m
KCl, 0.05
m
PO4, pH 6 buffer. Protein concentration was 15 μ
m
. Left side, bottom, a plot of the natural logarithm of the absorbance versus radial distance squared divided by 2 for the same data. The theoretical lines for a single species monomer (M), dimer (D), trimer (Tri), and tetramer (Tet) are shown for comparison. The data best fit a single species model with tetrameric molecular weight. Right side, middle, a plot of absorbance versus distance squared divided by 2 of the HR-C1 peptide at 22 °C and 30,000 rpm in a 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer. Peptide concentration was 250 μ
m
. Right side, bottom, a plot of the natural logarithm of the absorbance versus radial distance squared divided by 2 for the same data. The theoretical lines for monomer, dimer, and trimer are shown. The data were fit best to a monomer to trimer associating model in equilibrium. Residuals from both fits are shown above.
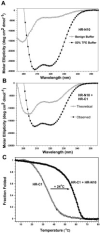
Fig. 6
Interaction between HR-N and HR-C peptides.A, circular dichroism spectra of HR-N10 peptide alone. Spectra were recorded at 25 °C in a 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer. For the spectrum containing 50% TFE, the above buffer was diluted 1:1 (v/v) with TFE. B, CD spectrum of a 1:1 molar complex between HR-N10 and HR-C1 peptides at 25 °C in a 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer. Peptide concentrations were 80 μ
m
. The theoretical spectrum for two noninteracting peptides is shown for comparison. This spectrum is generated by adding the individual peptide spectra at the same concentrations. C, temperature denaturation profiles of HR-C1 alone and a 1:1 molar HR-C1 with HR-N10 complex monitored by CD at 222 nm in a 0.1
m
KCl, 0.05
m
K2PO4, pH 7 buffer. Concentrations were 100 μ
m
. Fraction folded was calculated as described in Fig. 4 legend.
Fig. 7
HPLC analysis of the HR-N10/HR-C1 complex. HR-N10 (2 nmol) and HR-C1 (2 nmol) were pre-incubated together for 30 min in 10 μl of running buffer and then applied to a Superdex™ 75 SEC column equilibrated in a buffer consisting of 0.1
m
KCl, 0.05
m
PO4, pH 7, and a flow rate of 0.7 ml/min. The absorbance peak at 13 min corresponding to the HR-N10/HR-C1 complex was collected and subsequently analyzed by reversed phase chromatography (inset) on an analytical C8 Zorbax column employing a linear AB gradient of 2% B/min, where A = 0.05% aqueous trifluoroacetic acid and B is 0.05% trifluoroacetic acid/acetonitrile. Each absorbance peak is labeled accordingly.
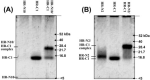
Fig. 8
Molecular mass of the complex formed between the HR-N and HR-C regions as determined by gel electrophoresis.A, HR-N10 and HR-C1 peptides on their own or as pre-incubated equimolar (200 μ
m
of each peptide) mixture were subjected to Tris/SDS-15% PAGE. Samples were incubated for 30 min in 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer, and the diluted 1:1 (v/v) with 2× Laemmli sample buffer at room temperature and loaded into the gel. The positions of HR-N10, HR-C1, and HR-N10/HR-C1 complex are indicated on the left side of the gel, whereas the positions of the molecule mass markers are indicated on the right side of the gels with arrows. Note the absence of the HR-N10 band was a result of the peptide eluting from the gel during the staining/destaining steps. B, HR-N2 and HR-C1 peptides treated in a manner similar to that described in A. The lanes of the gel, peptide locations, and molecular mass markers are labeled accordingly.
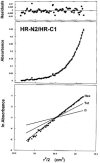
Fig. 9
Molecular mass of the complex formed between HR-N2 and HR-C1 peptides as determined by sedimentation equilibrium analysis at 22 °C and 30,000 rpm. The middle panel (absorbance versus radial distance squared divided by 2) shows the best fit curve for a single species model indicating a molecular mass of 30,500 Da. The theoretical mass for a 3:3 mole ratio HR-N2/HR-C1 hexamer is 31,692 Da. The upper panel shows the residuals from the curve fit, and the lower panel shows the ln absorbance _versus r_2/2 of the data compared with theoretical 1:1 dimer (D), 2:2 tetramer (Tet), and 3:3 hexamer (Hex).
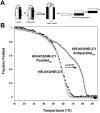
Fig. 10
Helix orientation in the HR-N/HR-C complex.A, schematic representation of the use of selective disulfide bridge formation to form parallel and anti-parallel hetero-stranded polypeptides, and the effect the relative helix orientations could have on the final folded state of the complex. The HR-N helix is represented as a white rectangle and corresponds to residues 916–950. The HR-C helix is represented as a black rectangle and corresponds to residues 1147–1185. The disulfide bridge between two cysteine residue side chains is denoted by S-S. B, temperature denaturation profiles of parallel and anti-parallel disulfide-bridged HR-N10/HR-C1 complexes in 0.1
m
KCl, 0.05
m
PO4, pH 7 buffer monitored by CD at 222 nm. Peptide concentrations were 15 μ
m
. Fraction folded was calculated as described in Fig. 4 legend. The temperature melting profile of the non-disulfide-bridged HR-N10/HR-C1 peptide is shown for comparison.
Similar articles
- Biophysical characterization of HRC peptide analogs interaction with heptad repeat regions of the SARS-coronavirus Spike fusion protein core.
Yan Z, Tripet B, Hodges RS. Yan Z, et al. J Struct Biol. 2006 Aug;155(2):162-75. doi: 10.1016/j.jsb.2006.03.024. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16765058 Free PMC article. - Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus.
Tripet B, Kao DJ, Jeffers SA, Holmes KV, Hodges RS. Tripet B, et al. J Struct Biol. 2006 Aug;155(2):176-94. doi: 10.1016/j.jsb.2006.03.019. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16697221 Free PMC article. - Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.
Xu Y, Zhu J, Liu Y, Lou Z, Yuan F, Liu Y, Cole DK, Ni L, Su N, Qin L, Li X, Bai Z, Bell JI, Pang H, Tien P, Gao GF, Rao Z. Xu Y, et al. Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555 - Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state.
Hakansson-McReynolds S, Jiang S, Rong L, Caffrey M. Hakansson-McReynolds S, et al. J Biol Chem. 2006 Apr 28;281(17):11965-71. doi: 10.1074/jbc.M601174200. Epub 2006 Feb 28. J Biol Chem. 2006. PMID: 16507566 Free PMC article. - The SARS-CoV S glycoprotein.
Xiao X, Dimitrov DS. Xiao X, et al. Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
Cited by
- Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms.
Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG. Adedeji AO, et al. J Virol. 2013 Jul;87(14):8017-28. doi: 10.1128/JVI.00998-13. Epub 2013 May 15. J Virol. 2013. PMID: 23678171 Free PMC article. - The S2 subunit of spike encodes diverse targets for functional antibody responses to SARS-CoV-2.
Guenthoer J, Garrett ME, Lilly M, Depierreux DM, Ruiz F, Chi M, Stoddard CI, Chohan V, Yaffe ZA, Sung K, Ralph D, Chu HY, Matsen FA 4th, Overbaugh J. Guenthoer J, et al. PLoS Pathog. 2024 Aug 2;20(8):e1012383. doi: 10.1371/journal.ppat.1012383. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39093891 Free PMC article. - Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein.
Deng Y, Liu J, Zheng Q, Yong W, Lu M. Deng Y, et al. Structure. 2006 May;14(5):889-99. doi: 10.1016/j.str.2006.03.007. Structure. 2006. PMID: 16698550 Free PMC article. - Biophysical characterization of HRC peptide analogs interaction with heptad repeat regions of the SARS-coronavirus Spike fusion protein core.
Yan Z, Tripet B, Hodges RS. Yan Z, et al. J Struct Biol. 2006 Aug;155(2):162-75. doi: 10.1016/j.jsb.2006.03.024. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16765058 Free PMC article. - Severe acute respiratory syndrome and coronavirus.
Hui DS, Chan PK. Hui DS, et al. Infect Dis Clin North Am. 2010 Sep;24(3):619-38. doi: 10.1016/j.idc.2010.04.009. Infect Dis Clin North Am. 2010. PMID: 20674795 Free PMC article. Review.
References
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous