A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans - PubMed (original) (raw)
A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans
Paula M Loria et al. J Neurosci. 2004.
Abstract
For a motor unit to function, neurons and muscle cells need to adopt their correct cell fate, form appropriate cellular contacts, and assemble a specific repertoire of signaling proteins into presynaptic and postsynaptic structures. In the nematode Caenorhabditis elegans, a disruption of any of these steps causes uncoordinated locomotory behavior (unc phenotype). We report here the positional cloning of a new unc gene, unc-122, which we show by mosaic analysis and tissue-specific rescue experiments to act in muscle to affect locomotory behavior. unc-122 codes for a phylogenetically conserved type II transmembrane protein with collagen repeats and a cysteine-rich olfactomedin domain. Together with uncharacterized proteins in C. elegans, Drosophila, and vertebrates, UNC-122 defines a novel family of proteins that we term "Colmedins." UNC-122 protein is expressed exclusively in muscle and coelomocytes and localizes to the postsynaptic surface of GABAergic and cholinergic neuromuscular junctions (NMJs). Presynaptic and postsynaptic structures are present and properly aligned in unc-122 mutant animals, yet the animals display neurotransmission defects characterized by an altered sensitivity toward drugs that interfere with cholinergic signaling. Moreover, unc-122 mutant animals display anatomical defects in motor axons that are likely a secondary consequence of neurotransmission defects. Both the neuroanatomical and locomotory defects worsen progressively during the life of an animal, consistent with a role of unc-122 in acute signaling at the NMJ. On the basis of motifs in the UNC-122 protein sequence that are characteristic of extracellular matrix proteins, we propose that UNC-122 is involved in maintaining a structural microenvironment that allows efficient neuromuscular signaling.
Figures
Figure 3.
UNC-122 is an OLF domain-containing protein. A, Map position of unc-122 and transformation rescue experiments. The approximate position of the eDf13 chromosomal deficiency is indicated as a gray bar. The right end extends to cosmid F11C3; the left end has not been mapped but must lie to the right of sur-2 (
). B, Schematic sequence of UNC-122 with an alignment of its conserved motifs and indication of location of mutant alleles. Residues that are conserved in >50% of aligned sequences are boxed. Note that the e2520 allele harbors two mutations. The ox79 and n2916 alleles, independently isolated in two different laboratories (see Materials and Methods), are identical mutations. ClustalX was used as the alignment tool (Thompson et al., 1997). Accession numbers for previously unpublished proteins are as follows: cof-2, AY494975; unc-122, AY494976; Gliomedin, NP852047.
Figure 6.
Reporter gene analysis of _cis-_regulatory elements of the unc-122 and cof-2 genes. A, unc-122 constructs used to generate transgenic lines. Black bars indicate genomic sequence (5′ and 3′ of the gene); the white box indicates the heterologous unc-54 3′UTR. Expression pattern and rescue of the unc-122 mutant phenotype is indicated (expression is shown in C). For localization of presumptive _cis-_regulatory regions driving expression in muscle, see Figure 5. GFP and HA epitope coding sequences are not drawn to scale. Antibody staining to detect the HA-tagged protein is shown in Figure 8 in the context of subcellular localization of UNC-122. B, cof-2 promoter construct. The promoter contains _cis-_regulatory elements for expression in coelomocytes and muscles as shown in _D_-G. _C_-G, Transgenic worms expressing gfp reporter gene constructs. Stars indicate coelomocytes; arrows indicate body wall muscles in E and enteric muscle in all other panels. Most body wall muscles are not visible in D because of mosaicism. C, unc-122prom::gfp (the 800 bp promoter fusion from A). _D_-G, cof-2prom::gfp.
Figure 5.
Functional dissection of the unc-122 locus: rescue analysis of fragments of the unc-122 locus. PCR products or subcloned DNA fragments were injected at 20 ng/μl into unc-122 mutant animals, and rescue of the coiling defect was scored. Rescue is defined as >50% of transgenic animals displaying wild-type (sinusoidal) movement patterns. Gray bars denote genomic regions 5′ and 3′ of the unc-122 locus. The deletion of the 5′ region without loss of rescuing activity argues for the presence of cryptic internal transcriptional and translational start sites. Potential alternative ATG start codons followed by cryptic potential signal sequences are indicated with gray arrows. Regions corresponding to the transmembrane domain (black), collagen repeats (blue), and OLF domain (red) are in color.
Figure 1.
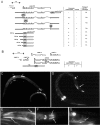
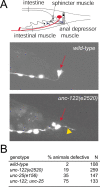
Motor axon anatomy defects of unc-122 mutant animals. A, Ectopic axon outgrowth (yellow arrowhead) from the DVB motor neuron (red arrow points to cell body), visualized with the unc-47::gfp reporter transgene oxIs12 in an adult animal. A schematic drawing of DVB motor neuron morphology is shown in the top panel, with synapses to its enteric muscle targets labeled schematically (blue arrows). B, Quantification of axon sprouting defects. Animals were raised at 25°C and scored as adults.
Figure 2.
Locomotory defects of unc-122(e2520) mutant animals. A, Body bends per time interval of adult animals moving on an agar plate at room temperature. B, Thrashing of adult animals at room temperature in a drop of M9 buffer. C, Reversal behavior of adult animals at room temperature on a nonseeded agar plate. D, Aldicarb (1 m
m
)-induced paralysis of adult animals cultivated at 20°C. E, Levamisole (100 m
m
)-induced paralysis of adult animals cultivated at 20°C.
Figure 4.
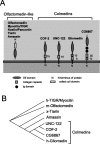
UNC-122 is phylogenetically conserved. A, Protein domain structure of OLF domain-containing proteins. Vertebrate genomes contain one other OLF domain-containing protein type, Latrophilin, a protein with various extracellular domains and seven transmembrane domains (Lelianova et al., 1997). Worms and flies each contain a single ortholog of Latrophilin that, however, contains no OLF domain. We named the family of collagen repeat-plus olfactomedin domain-containing proteins the Colmedins. B, Phylogenetic analysis of the OLF domain. A cladogram of a neighbor-joining tree was constructed using distance analysis in the PAUP program package (Swofford, 2000).
Figure 7.
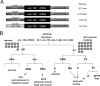
unc-122 acts in muscle. A, Tissue-specific rescue experiments. Constructs (not drawn to scale) were injected into unc-122(e2520) animals at 20 ng/μl. Rescue was scored as a line having >50% of animals that locomote without coiling. B, Mosaic analysis. The 5 kb unc-122 rescuing fragment (Fig. 2_A_) was injected into unc-122(e2520) animals (at 20 ng/μl) together with ttx-3::gfp and myo-3::gfp as lineage markers (at 50 ng/μl). Two classes of animals were scored for rescue of locomotory defects. First, animals in which AB cell descendants (_ttx-3::gfp_-expressing AIY cells and _myo-3::gfp_-expressing enteric muscle cells) express gfp but not P cell descendents (body wall muscle); second, animals in which body wall muscle, but not AIY or enteric muscles, expresses gfp. Gray circles indicate how many animals per class were found that had lost the array and were scored for rescue.
Figure 8.
UNC-122 is enriched at muscle postsynaptic zones but does not affect their development. A, Schematic drawing of individual muscle cells and their muscle arms that extend into the VNC to make neuromuscular junctions (circled) with individual motor axons (red). Only a subset of cells and NMJs are shown. Note that individual motor neurons make multiple en passant synapses with many muscle arms and that muscle arm and endplate morphology can be very complex, stretching over a substantial area; this is shown in more detail in supplemental Figure 1. B, UNC-122 is enriched at NMJs. All animals contain an _unc-122::HA-_expressing, chromosomally integrated array (otIs127) that rescues the mutant unc-122 phenotype. To visualize UNC-38 localization, the ljEx42 array was used. Colocalization of UNC-122 to muscle endplates was observed not only with UNC-38 but also with UNC-29 (data not shown). In unc-6 mutants, neurons and consequently muscle arms are mislocalized, causing the generation of aberrantly localized NMJs. C, Presynaptic and postsynaptic differentiation of NMJs appears normal in unc-122 mutants. All panels show unc-122 mutant animals. In the top panel, cholinergic presynaptic specialization (visualized with anti-UNC-17) and postsynaptic GABAergic NMJs (visualized with antibody staining against UNC-49::GFP; note the visualization and intactness of muscle arms) are shown; in the bottom panel, presynaptic and postsynaptic cholinergic specializations are shown. The animals in the bottom panel contain the unc-29(x568) allele in the background, which is necessary for correct localization of the unc-29::gfp reporter expressed from the xEx1 array (Fleming et al., 1997). Wild-type controls appear indistinguishable (data not shown).
Similar articles
- Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission.
Loria PM, Duke A, Rand JB, Hobert O. Loria PM, et al. Curr Biol. 2003 Aug 5;13(15):1317-23. doi: 10.1016/s0960-9822(03)00532-3. Curr Biol. 2003. PMID: 12906792 - Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system.
Starich TA, Xu J, Skerrett IM, Nicholson BJ, Shaw JE. Starich TA, et al. Neural Dev. 2009 May 11;4:16. doi: 10.1186/1749-8104-4-16. Neural Dev. 2009. PMID: 19432959 Free PMC article. - Localization mechanisms of the axon guidance molecule UNC-6/Netrin and its receptors, UNC-5 and UNC-40, in Caenorhabditis elegans.
Ogura K, Asakura T, Goshima Y. Ogura K, et al. Dev Growth Differ. 2012 Apr;54(3):390-7. doi: 10.1111/j.1440-169X.2012.01349.x. Dev Growth Differ. 2012. PMID: 22524608 Review. - C. elegans Locomotion: Finding Balance in Imbalance.
Thapliyal S, Babu K. Thapliyal S, et al. Adv Exp Med Biol. 2018;1112:185-196. doi: 10.1007/978-981-13-3065-0_14. Adv Exp Med Biol. 2018. PMID: 30637699 Review.
Cited by
- Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system.
Toth ML, Melentijevic I, Shah L, Bhatia A, Lu K, Talwar A, Naji H, Ibanez-Ventoso C, Ghose P, Jevince A, Xue J, Herndon LA, Bhanot G, Rongo C, Hall DH, Driscoll M. Toth ML, et al. J Neurosci. 2012 Jun 27;32(26):8778-90. doi: 10.1523/JNEUROSCI.1494-11.2012. J Neurosci. 2012. PMID: 22745480 Free PMC article. - The FoxF/FoxC factor LET-381 directly regulates both cell fate specification and cell differentiation in C. elegans mesoderm development.
Amin NM, Shi H, Liu J. Amin NM, et al. Development. 2010 May;137(9):1451-60. doi: 10.1242/dev.048496. Epub 2010 Mar 24. Development. 2010. PMID: 20335356 Free PMC article. - The genomic environment around the Aromatase gene: evolutionary insights.
Castro LF, Santos MM, Reis-Henriques MA. Castro LF, et al. BMC Evol Biol. 2005 Aug 12;5:43. doi: 10.1186/1471-2148-5-43. BMC Evol Biol. 2005. PMID: 16098224 Free PMC article. - Imaging Glycosaminoglycan Modification Patterns In Vivo.
Bülow HE. Bülow HE. Methods Mol Biol. 2022;2303:539-557. doi: 10.1007/978-1-0716-1398-6_42. Methods Mol Biol. 2022. PMID: 34626406 - Measuring Caenorhabditis elegans Sensitivity to the Acetylcholine Receptor Agonist Levamisole.
Davis AN, Tanis JE. Davis AN, et al. J Vis Exp. 2022 Jun 7;(184):10.3791/64056. doi: 10.3791/64056. J Vis Exp. 2022. PMID: 35758705 Free PMC article.
References
- Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB (1993) The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261: 617-619. - PubMed
- Avery L, Thomas JH (1997) Feeding and defecation. In: C. elegans II (Riddle DL, Blumenthal T, Meyer BJ, Priess JR, eds), pp 679-716. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. - PubMed
- Brown MC, Holland RL, Hopkins WG (1981) Motor nerve sprouting. Annu Rev Neurosci 4: 17-42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases